

You can:
| Name | C-C chemokine receptor type 5 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | CCR5 |
| Synonym | CD195 chemokine (C-C motif) receptor 5 (gene/pseudogene) CHEMR13 CCR5 CCR-5 [ Show all ] |
| Disease | Human immunodeficiency virus infection |
| Length | 352 |
| Amino acid sequence | MDYQVSSPIYDINYYTSEPCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINCKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPGIIFTRSQKEGLHYTCSSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRCRNEKKRHRAVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEKFRNYLLVFFQKHIAKRFCKCCSIFQQEAPERASSVYTRSTGEQEISVGL |
| UniProt | P51681 |
| Protein Data Bank | 4mbs, 6aky, 6akx, 5uiw |
| GPCR-HGmod model | P51681 |
| 3D structure model | This structure is from PDB ID 4mbs. |
| BioLiP | BL0430746, BL0430745, BL0430742,BL0430744, BL0430741,BL0430743, BL0382816, BL0256313,BL0256315, BL0256312,BL0256314 |
| Therapeutic Target Database | T72171 |
| ChEMBL | CHEMBL274 |
| IUPHAR | 62 |
| DrugBank | BE0000911 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 132 | 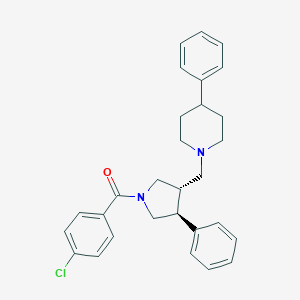 CHEMBL316073 CHEMBL316073 | C29H31ClN2O | 459.03 | 2 / 0 | 6.1 | No |
| 164 | 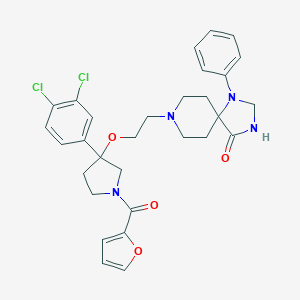 CHEMBL522158 CHEMBL522158 | C30H32Cl2N4O4 | 583.51 | 6 / 1 | 4.8 | No |
| 170 | 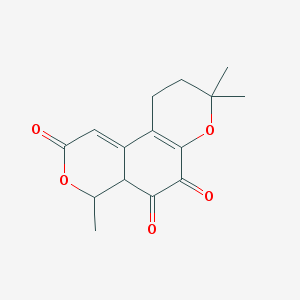 BDBM50242234 BDBM50242234 | C15H16O5 | 276.288 | 5 / 0 | 0.7 | Yes |
| 360 | 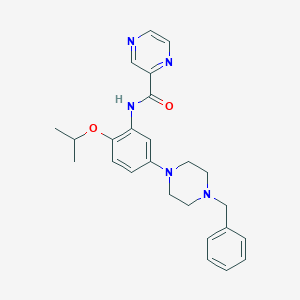 CHEMBL3263023 CHEMBL3263023 | C25H29N5O2 | 431.54 | 6 / 1 | 3.1 | Yes |
| 386 | 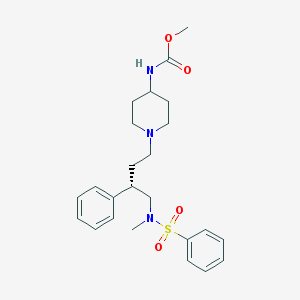 CHEMBL310369 CHEMBL310369 | C24H33N3O4S | 459.605 | 6 / 1 | 3.5 | Yes |
| 455 | 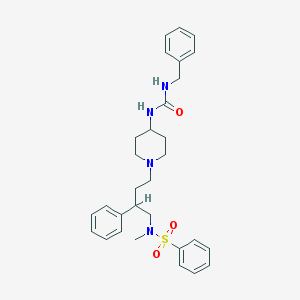 CHEMBL309922 CHEMBL309922 | C30H38N4O3S | 534.719 | 5 / 2 | 4.4 | No |
| 578 | 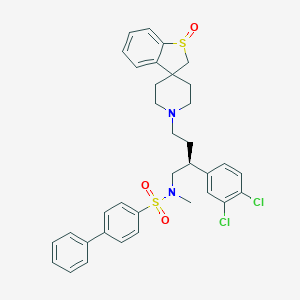 CHEMBL330570 CHEMBL330570 | C35H36Cl2N2O3S2 | 667.704 | 6 / 0 | 7.2 | No |
| 711 | 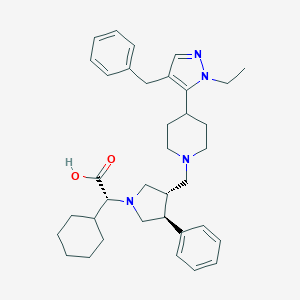 CHEMBL368265 CHEMBL368265 | C36H48N4O2 | 568.806 | 5 / 1 | 4.6 | No |
| 441695 | 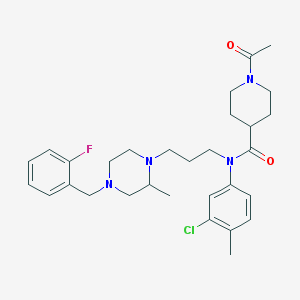 CHEMBL3397986 CHEMBL3397986 | C30H40ClFN4O2 | 543.124 | 5 / 0 | 4.5 | No |
| 728 | 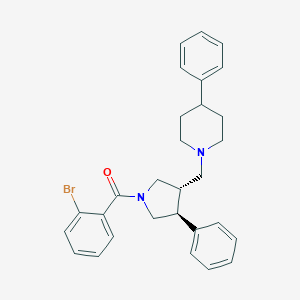 CHEMBL93168 CHEMBL93168 | C29H31BrN2O | 503.484 | 2 / 0 | 6.1 | No |
| 756 | 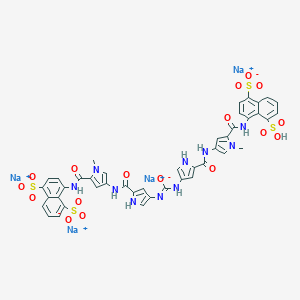 CID 136859397 CID 136859397 | C43H32N10Na4O17S4 | 1180.98 | 18 / 8 | N/A | No |
| 779 | 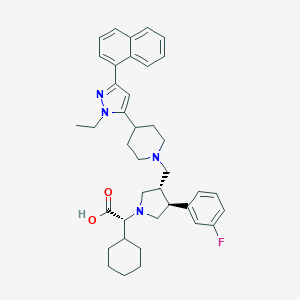 CHEMBL172944 CHEMBL172944 | C39H47FN4O2 | 622.829 | 6 / 1 | 5.7 | No |
| 1079 | 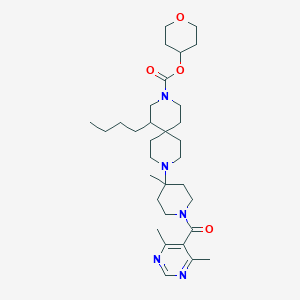 CHEMBL499729 CHEMBL499729 | C32H51N5O4 | 569.791 | 7 / 0 | 4.5 | No |
| 1080 | 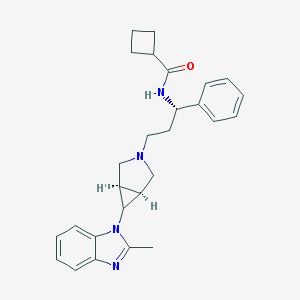 CHEMBL2203618 CHEMBL2203618 | C27H32N4O | 428.58 | 3 / 1 | 4.1 | Yes |
| 1695 | 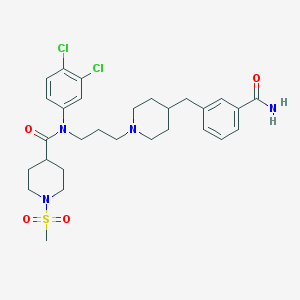 CHEMBL382252 CHEMBL382252 | C29H38Cl2N4O4S | 609.607 | 6 / 1 | 4.3 | No |
| 1705 | 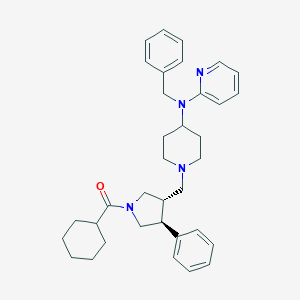 AC1LA24O AC1LA24O | C35H44N4O | 536.764 | 4 / 0 | 6.8 | No |
| 1800 | 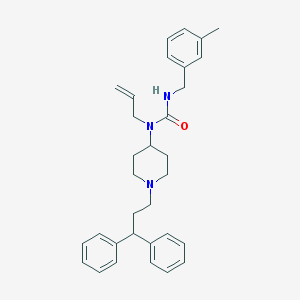 CHEMBL183261 CHEMBL183261 | C32H39N3O | 481.684 | 2 / 1 | 6.4 | No |
| 2235 | 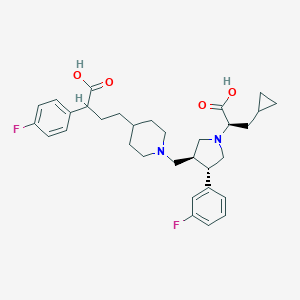 CHEMBL284010 CHEMBL284010 | C32H40F2N2O4 | 554.679 | 8 / 2 | 1.7 | No |
| 2328 | 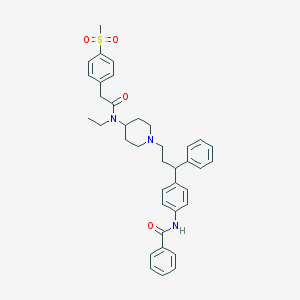 CHEMBL199119 CHEMBL199119 | C38H43N3O4S | 637.839 | 5 / 1 | 6.1 | No |
| 3202 | 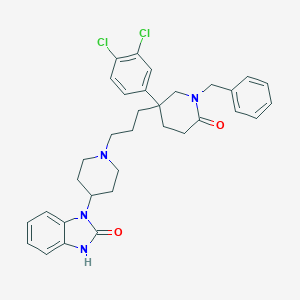 CHEMBL26790 CHEMBL26790 | C33H36Cl2N4O2 | 591.577 | 3 / 1 | 6.1 | No |
| 3502 | 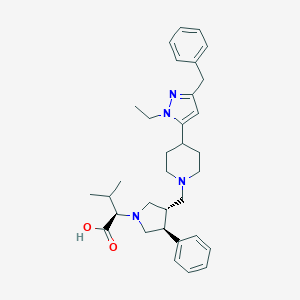 CHEMBL172551 CHEMBL172551 | C33H44N4O2 | 528.741 | 5 / 1 | 3.6 | No |
| 3587 | 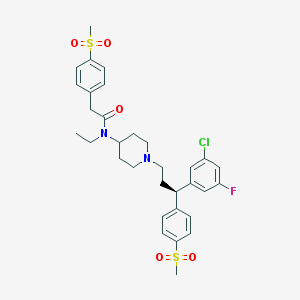 CHEMBL382435 CHEMBL382435 | C32H38ClFN2O5S2 | 649.233 | 7 / 0 | 5.2 | No |
| 3824 | 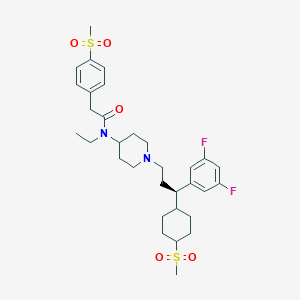 CHEMBL2178577 CHEMBL2178577 | C32H44F2N2O5S2 | 638.83 | 8 / 0 | 4.7 | No |
| 3826 | 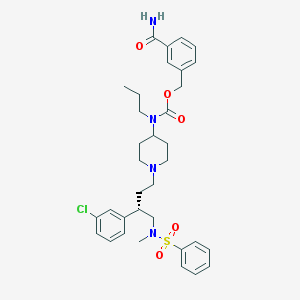 CHEMBL310106 CHEMBL310106 | C34H43ClN4O5S | 655.251 | 7 / 1 | 5.6 | No |
| 4229 | 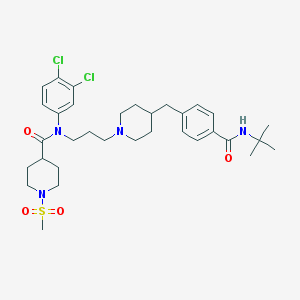 CHEMBL205964 CHEMBL205964 | C33H46Cl2N4O4S | 665.715 | 6 / 1 | 5.6 | No |
| 4812 | 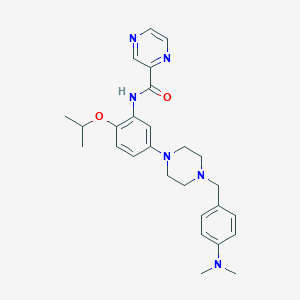 CHEMBL3263322 CHEMBL3263322 | C27H34N6O2 | 474.609 | 7 / 1 | 3.3 | Yes |
| 4848 | 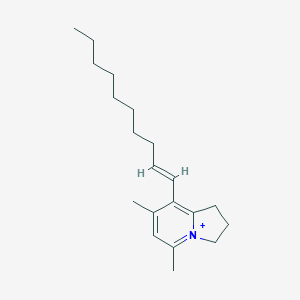 CHEMBL2064265 CHEMBL2064265 | C20H32N+ | 286.483 | 0 / 0 | 6.8 | No |
| 4849 | 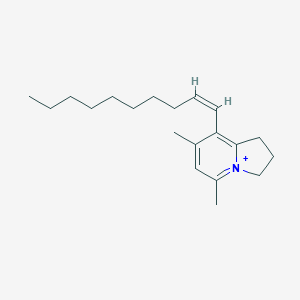 CHEMBL2057534 CHEMBL2057534 | C20H32N+ | 286.483 | 0 / 0 | 6.8 | No |
| 5471 | 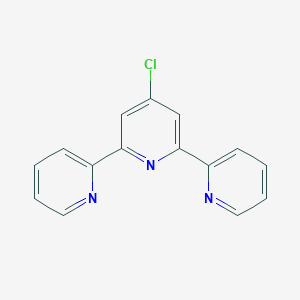 4'-Chloro-2,2':6',2''-terpyridine 4'-Chloro-2,2':6',2''-terpyridine | C15H10ClN3 | 267.716 | 3 / 0 | 3.0 | Yes |
| 5592 | 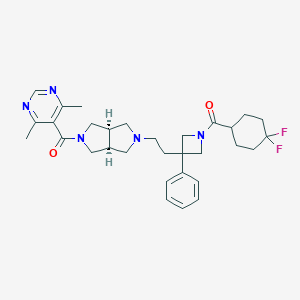 CHEMBL592940 CHEMBL592940 | C31H39F2N5O2 | 551.683 | 7 / 0 | 3.8 | No |
| 5888 | 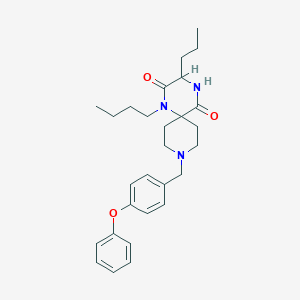 CHEMBL1171291 CHEMBL1171291 | C28H37N3O3 | 463.622 | 4 / 1 | 4.8 | Yes |
| 6092 | 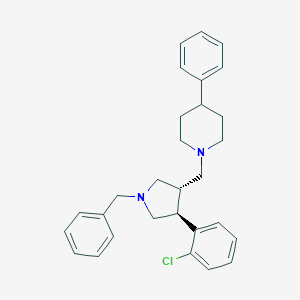 CHEMBL327478 CHEMBL327478 | C29H33ClN2 | 445.047 | 2 / 0 | 6.4 | No |
| 6149 | 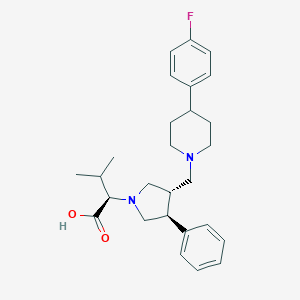 CHEMBL322693 CHEMBL322693 | C27H35FN2O2 | 438.587 | 5 / 1 | 3.1 | Yes |
| 6663 | 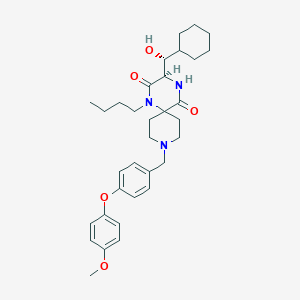 CHEMBL1813269 CHEMBL1813269 | C33H45N3O5 | 563.739 | 6 / 2 | 5.8 | No |
| 7097 | 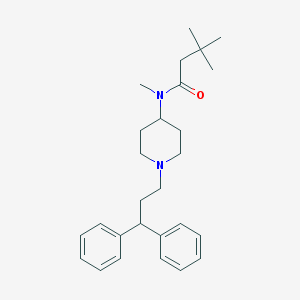 CHEMBL427137 CHEMBL427137 | C27H38N2O | 406.614 | 2 / 0 | 5.7 | No |
| 7299 | 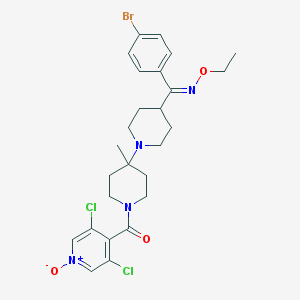 CHEMBL164484 CHEMBL164484 | C26H31BrCl2N4O3 | 598.363 | 5 / 0 | 4.8 | No |
| 7300 | 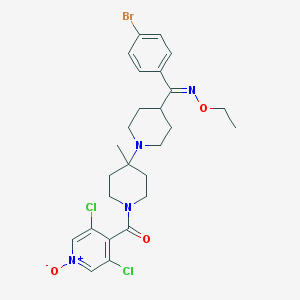 CHEMBL138640 CHEMBL138640 | C26H31BrCl2N4O3 | 598.363 | 5 / 0 | 4.8 | No |
| 7445 | 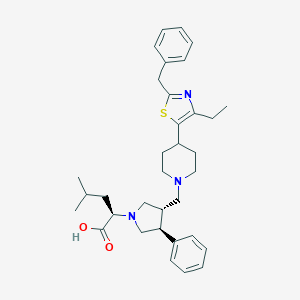 CHEMBL480890 CHEMBL480890 | C34H45N3O2S | 559.813 | 6 / 1 | 5.2 | No |
| 7470 | 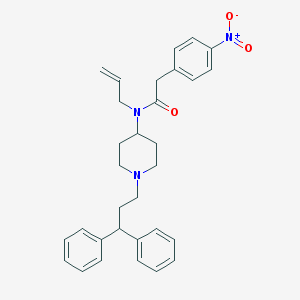 CHEMBL426960 CHEMBL426960 | C31H35N3O3 | 497.639 | 4 / 0 | 6.1 | No |
| 7490 | 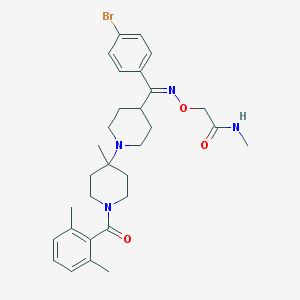 CHEMBL341928 CHEMBL341928 | C30H39BrN4O3 | 583.571 | 5 / 1 | 5.3 | No |
| 7529 | 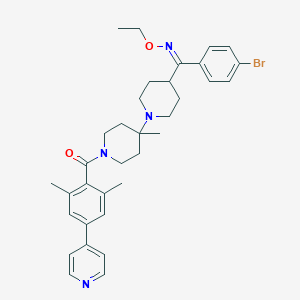 CHEMBL140484 CHEMBL140484 | C34H41BrN4O2 | 617.632 | 5 / 0 | 6.9 | No |
| 7699 | 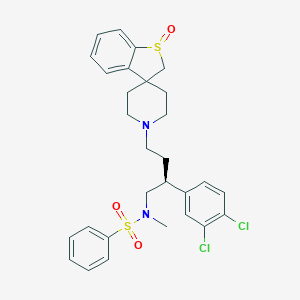 CHEMBL79504 CHEMBL79504 | C29H32Cl2N2O3S2 | 591.606 | 6 / 0 | 5.5 | No |
| 7924 | 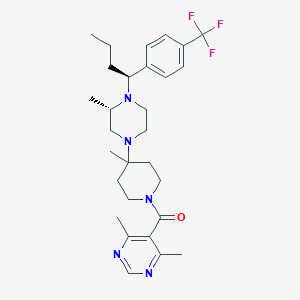 CHEMBL432170 CHEMBL432170 | C29H40F3N5O | 531.668 | 8 / 0 | 5.1 | No |
| 8160 | 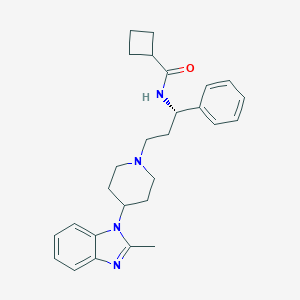 CHEMBL2203614 CHEMBL2203614 | C27H34N4O | 430.596 | 3 / 1 | 4.5 | Yes |
| 8401 | 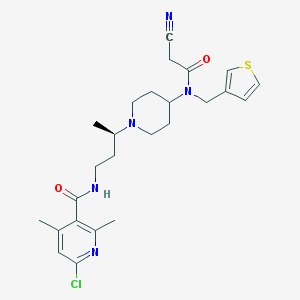 CHEMBL2164220 CHEMBL2164220 | C25H32ClN5O2S | 502.074 | 6 / 1 | 3.8 | No |
| 8425 | 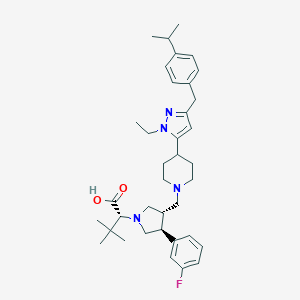 CHEMBL263529 CHEMBL263529 | C37H51FN4O2 | 602.839 | 6 / 1 | 5.2 | No |
| 8612 | 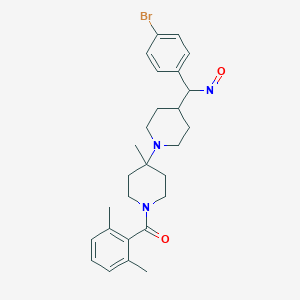 BDBM50115530 BDBM50115530 | C27H34BrN3O2 | 512.492 | 4 / 0 | 5.4 | No |
| 8789 | 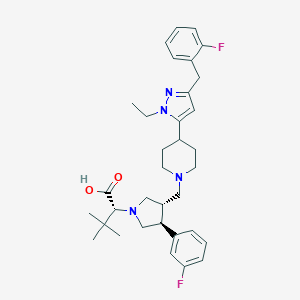 CHEMBL177071 CHEMBL177071 | C34H44F2N4O2 | 578.749 | 7 / 1 | 4.2 | No |
| 9280 | 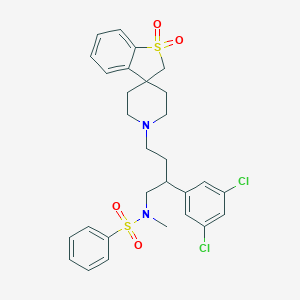 CHEMBL91374 CHEMBL91374 | C29H32Cl2N2O4S2 | 607.605 | 6 / 0 | 5.7 | No |
| 9283 | 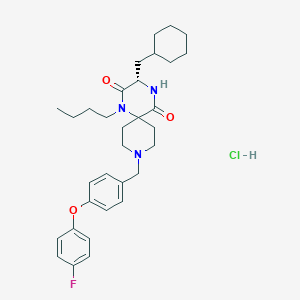 CHEMBL1173647 CHEMBL1173647 | C32H43ClFN3O3 | 572.162 | 5 / 2 | N/A | No |
| 9299 | 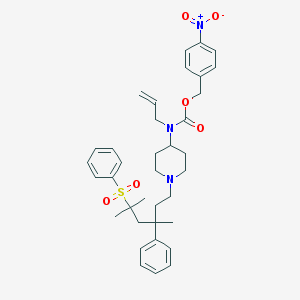 CHEMBL118544 CHEMBL118544 | C36H45N3O6S | 647.831 | 7 / 0 | 7.2 | No |
| 9768 | 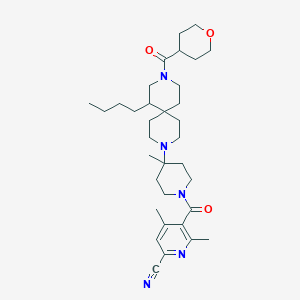 CHEMBL505595 CHEMBL505595 | C34H51N5O3 | 577.814 | 6 / 0 | 4.7 | No |
| 9978 | 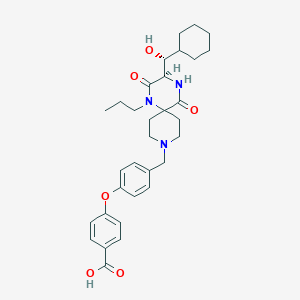 CHEMBL1813448 CHEMBL1813448 | C32H41N3O6 | 563.695 | 7 / 3 | 2.8 | No |
| 442094 | 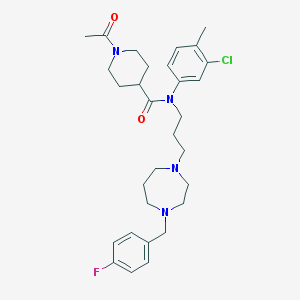 CHEMBL3397998 CHEMBL3397998 | C30H40ClFN4O2 | 543.124 | 5 / 0 | 4.4 | No |
| 10453 | 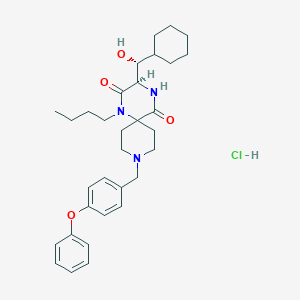 CHEMBL536754 CHEMBL536754 | C32H44ClN3O4 | 570.171 | 5 / 3 | N/A | No |
| 10454 | 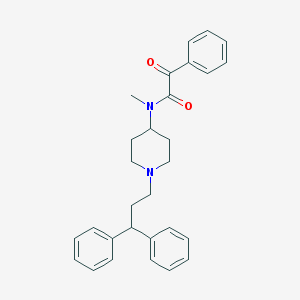 CHEMBL182403 CHEMBL182403 | C29H32N2O2 | 440.587 | 3 / 0 | 5.6 | No |
| 10503 | 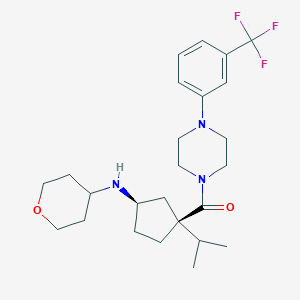 CHEMBL1683055 CHEMBL1683055 | C25H36F3N3O2 | 467.577 | 7 / 1 | 4.2 | Yes |
| 10640 | 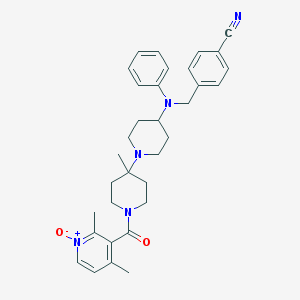 CHEMBL58248 CHEMBL58248 | C33H39N5O2 | 537.708 | 5 / 0 | 4.4 | No |
| 10691 | 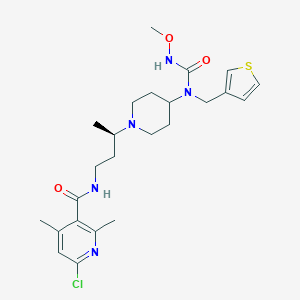 CHEMBL2164206 CHEMBL2164206 | C24H34ClN5O3S | 508.078 | 6 / 2 | 3.8 | No |
| 11360 | 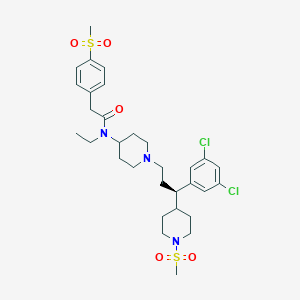 CHEMBL1952100 CHEMBL1952100 | C31H43Cl2N3O5S2 | 672.721 | 7 / 0 | 4.9 | No |
| 11382 | 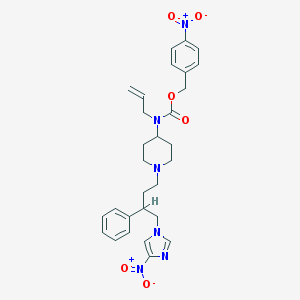 CHEMBL322439 CHEMBL322439 | C29H34N6O6 | 562.627 | 8 / 0 | 4.7 | No |
| 11469 | 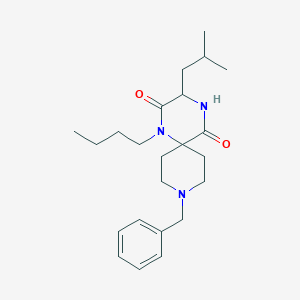 SCHEMBL6012808 SCHEMBL6012808 | C23H35N3O2 | 385.552 | 3 / 1 | 3.7 | Yes |
| 11474 | 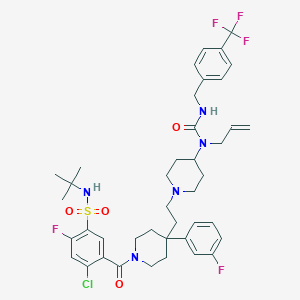 CHEMBL1288880 CHEMBL1288880 | C41H49ClF5N5O4S | 838.376 | 11 / 2 | 7.7 | No |
| 11476 | 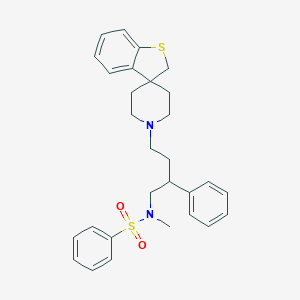 CHEMBL88455 CHEMBL88455 | C29H34N2O2S2 | 506.723 | 5 / 0 | 5.7 | No |
| 11870 |  CHEMBL205851 CHEMBL205851 | C28H35Cl2FN4O2 | 549.512 | 5 / 1 | 5.2 | No |
| 464309 |  CHEMBL3604302 CHEMBL3604302 | C26H26Cl3N3O2 | 518.863 | 3 / 2 | N/A | No |
| 12072 |  CHEMBL182557 CHEMBL182557 | C29H33FN2O | 444.594 | 3 / 0 | 5.8 | No |
| 12175 |  SCHEMBL3459521 SCHEMBL3459521 | C40H51N5O4S | 697.939 | 8 / 1 | 6.9 | No |
| 12176 | 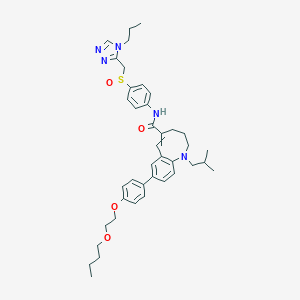 AQXIPTPSJDOCRP-UHFFFAOYSA-N AQXIPTPSJDOCRP-UHFFFAOYSA-N | C40H51N5O4S | 697.939 | 8 / 1 | 6.9 | No |
| 12229 | 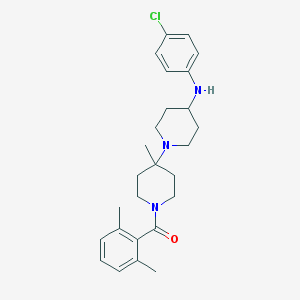 CHEMBL61466 CHEMBL61466 | C26H34ClN3O | 440.028 | 3 / 1 | 5.7 | No |
| 12412 | 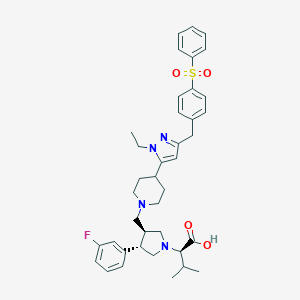 CHEMBL177156 CHEMBL177156 | C39H47FN4O4S | 686.887 | 8 / 1 | 4.5 | No |
| 13190 | 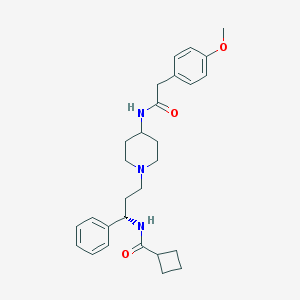 CHEMBL460630 CHEMBL460630 | C28H37N3O3 | 463.622 | 4 / 2 | 3.9 | Yes |
| 13343 | 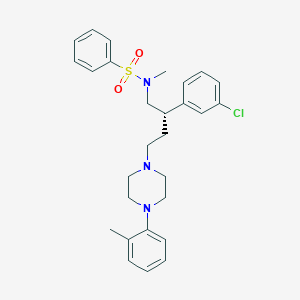 CHEMBL79369 CHEMBL79369 | C28H34ClN3O2S | 512.109 | 5 / 0 | 5.8 | No |
| 13453 | 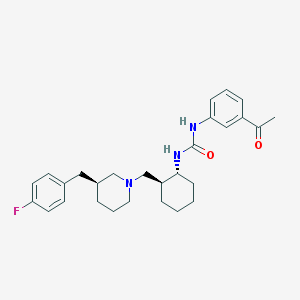 CHEMBL250689 CHEMBL250689 | C28H36FN3O2 | 465.613 | 4 / 2 | 5.2 | No |
| 13479 | 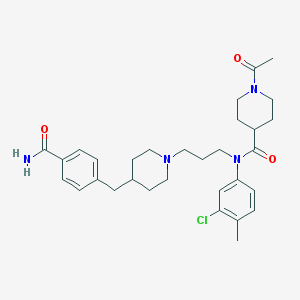 TAK-220 TAK-220 | C31H41ClN4O3 | 553.144 | 4 / 1 | 4.2 | No |
| 13547 | 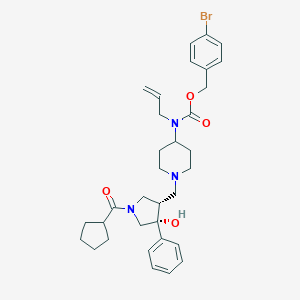 CHEMBL1172044 CHEMBL1172044 | C33H42BrN3O4 | 624.62 | 5 / 1 | 5.3 | No |
| 13752 | 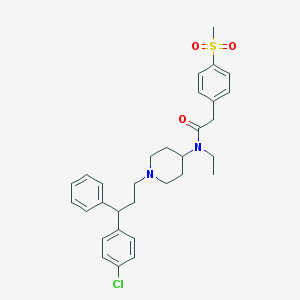 CHEMBL198277 CHEMBL198277 | C31H37ClN2O3S | 553.158 | 4 / 0 | 5.9 | No |
| 13948 | 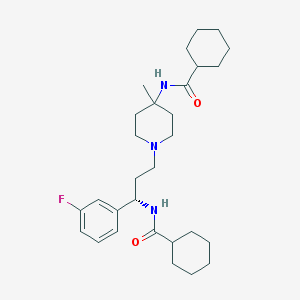 CHEMBL1171877 CHEMBL1171877 | C29H44FN3O2 | 485.688 | 4 / 2 | 5.8 | No |
| 13963 | 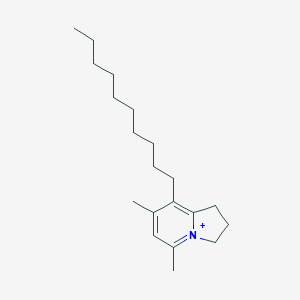 CHEMBL2064266 CHEMBL2064266 | C20H34N+ | 288.499 | 0 / 0 | 7.2 | No |
| 14266 |  CHEMBL161093 CHEMBL161093 | C33H40N2O | 480.696 | 3 / 0 | 7.6 | No |
| 14411 | 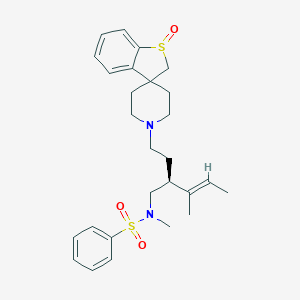 CHEMBL2259883 CHEMBL2259883 | C27H36N2O3S2 | 500.716 | 6 / 0 | 4.3 | No |
| 14415 | 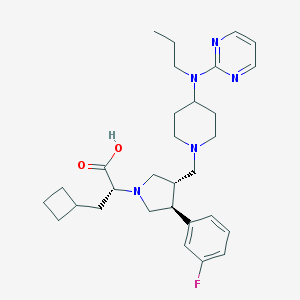 CHEMBL428104 CHEMBL428104 | C30H42FN5O2 | 523.697 | 8 / 1 | 3.4 | No |
| 15095 | 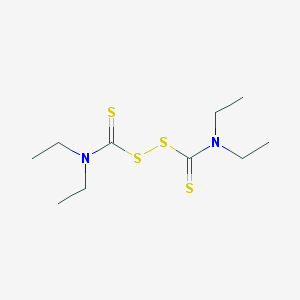 disulfiram disulfiram | C10H20N2S4 | 296.524 | 4 / 0 | 3.9 | Yes |
| 16015 | 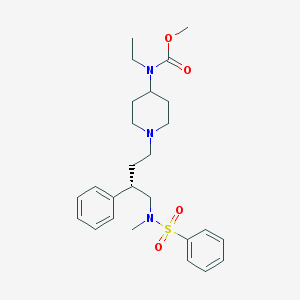 CHEMBL310645 CHEMBL310645 | C26H37N3O4S | 487.659 | 6 / 0 | 4.0 | Yes |
| 16405 | 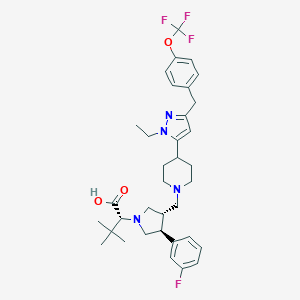 CHEMBL176426 CHEMBL176426 | C35H44F4N4O3 | 644.756 | 10 / 1 | 5.3 | No |
| 16611 | 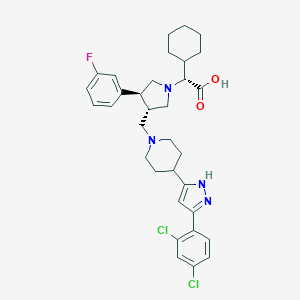 CHEMBL170717 CHEMBL170717 | C33H39Cl2FN4O2 | 613.599 | 6 / 2 | 5.4 | No |
| 16847 | 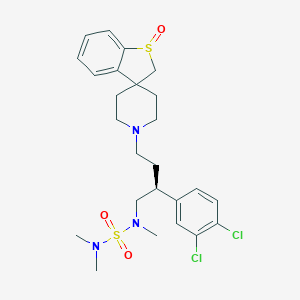 CHEMBL90272 CHEMBL90272 | C25H33Cl2N3O3S2 | 558.577 | 7 / 0 | 3.9 | No |
| 17306 | 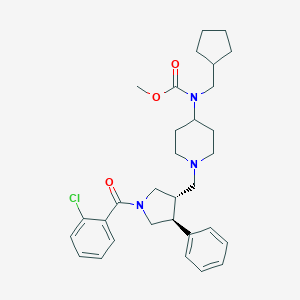 CHEMBL93961 CHEMBL93961 | C31H40ClN3O3 | 538.129 | 4 / 0 | 6.1 | No |
| 17479 | 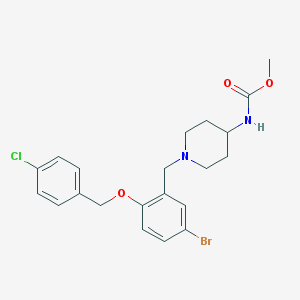 CHEMBL234843 CHEMBL234843 | C21H24BrClN2O3 | 467.788 | 4 / 1 | 4.8 | Yes |
| 17696 | 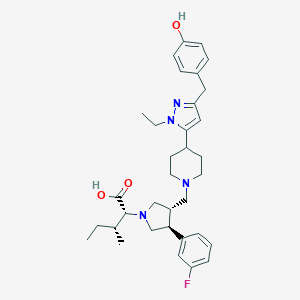 CHEMBL408800 CHEMBL408800 | C34H45FN4O3 | 576.757 | 7 / 2 | 3.7 | No |
| 17697 | 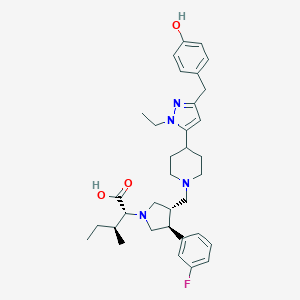 CHEMBL368375 CHEMBL368375 | C34H45FN4O3 | 576.757 | 7 / 2 | 3.7 | No |
| 17805 | 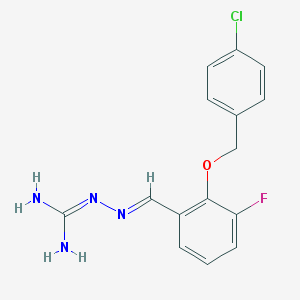 CHEMBL395844 CHEMBL395844 | C15H14ClFN4O | 320.752 | 4 / 2 | 2.7 | Yes |
| 17819 | 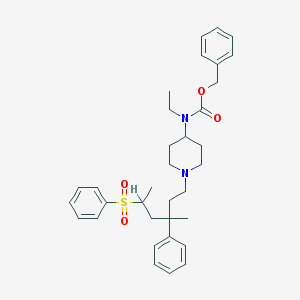 CHEMBL118196 CHEMBL118196 | C34H44N2O4S | 576.796 | 5 / 0 | 7.0 | No |
| 17864 | 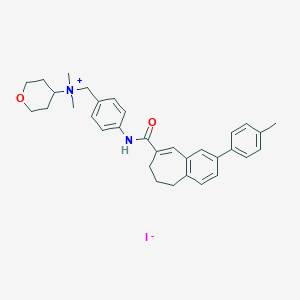 CHEMBL64322 CHEMBL64322 | C33H39IN2O2 | 622.591 | 3 / 1 | N/A | No |
| 18032 | 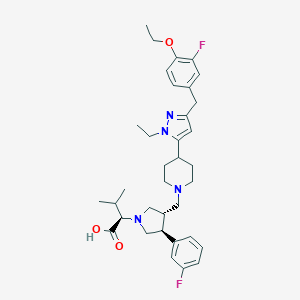 CHEMBL175348 CHEMBL175348 | C35H46F2N4O3 | 608.775 | 8 / 1 | 4.1 | No |
| 18254 | 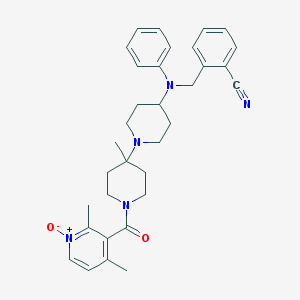 CHEMBL418120 CHEMBL418120 | C33H39N5O2 | 537.708 | 5 / 0 | 4.4 | No |
| 18403 | 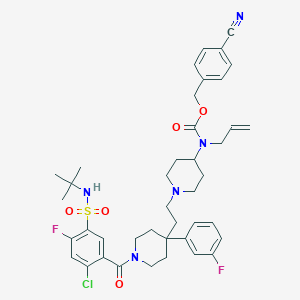 CHEMBL1288877 CHEMBL1288877 | C41H48ClF2N5O5S | 796.372 | 10 / 1 | 7.1 | No |
| 18450 | 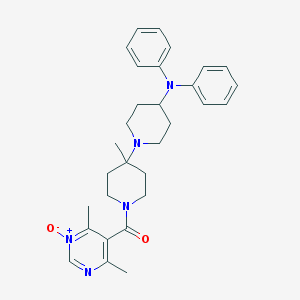 CHEMBL58004 CHEMBL58004 | C30H37N5O2 | 499.659 | 5 / 0 | 4.2 | Yes |
| 465164 | 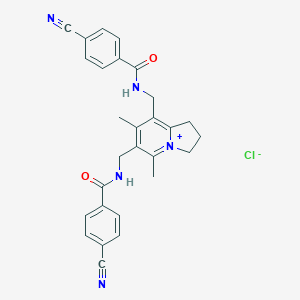 CHEMBL3604306 CHEMBL3604306 | C28H26ClN5O2 | 499.999 | 5 / 2 | N/A | N/A |
| 18963 | 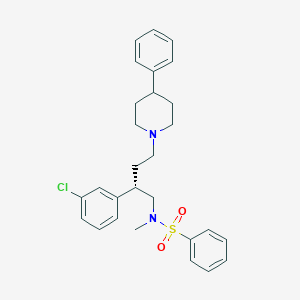 CHEMBL312701 CHEMBL312701 | C28H33ClN2O2S | 497.094 | 4 / 0 | 6.1 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417