

You can:
| Name | D(2) dopamine receptor |
|---|---|
| Species | Mus musculus (Mouse) |
| Gene | Drd2 |
| Synonym | D2 receptor D2(415) and D2(444) D2A and D2B D2R dopamine D2 receptor [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 444 |
| Amino acid sequence | MDPLNLSWYDDDLERQNWSRPFNGSEGKPDRPHYNYYAMLLTLLIFIIVFGNVLVCMAVSREKALQTTTNYLIVSLAVADLLVATLVMPWVVYLEVVGEWKFSRIHCDIFVTLDVMMCTASILNLCAISIDRYTAVAMPMLYNTRYSSKRRVTVMIAIVWVLSFTISCPLLFGLNNTDQNECIIANPAFVVYSSIVSFYVPFIVTLLVYIKIYIVLRKRRKRVNTKRSSRAFRANLKTPLKGNCTHPEDMKLCTVIMKSNGSFPVNRRRMDAARRAQELEMEMLSSTSPPERTRYSPIPPSHHQLTLPDPSHHGLHSNPDSPAKPEKNGHAKIVNPRIAKFFEIQTMPNGKTRTSLKTMSRRKLSQQKEKKATQMLAIVLGVFIICWLPFFITHILNIHCDCNIPPVLYSAFTWLGYVNSAVNPIIYTTFNIEFRKAFMKILHC |
| UniProt | P61168 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL3427 |
| IUPHAR | 215 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 269 | 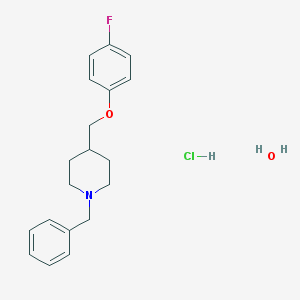 CHEMBL539358 CHEMBL539358 | C19H25ClFNO2 | 353.862 | 4 / 2 | N/A | N/A |
| 4231 | 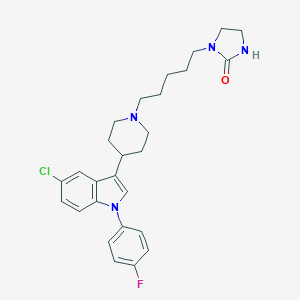 CHEMBL418160 CHEMBL418160 | C27H32ClFN4O | 483.028 | 3 / 1 | 5.1 | No |
| 4544 | 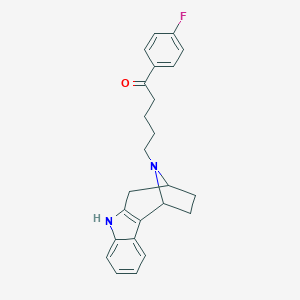 CHEMBL285712 CHEMBL285712 | C24H25FN2O | 376.475 | 3 / 1 | 4.7 | Yes |
| 459266 | 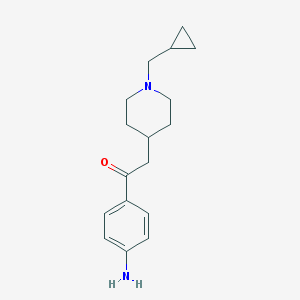 CHEMBL137163 CHEMBL137163 | C17H24N2O | 272.392 | 3 / 1 | 2.4 | Yes |
| 6825 | 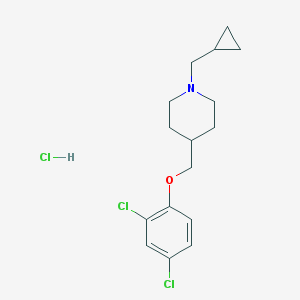 CHEMBL540322 CHEMBL540322 | C16H22Cl3NO | 350.708 | 2 / 1 | N/A | N/A |
| 6869 | 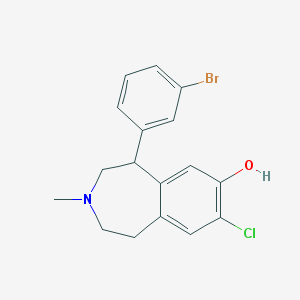 CHEMBL13632 CHEMBL13632 | C17H17BrClNO | 366.683 | 2 / 1 | 4.7 | Yes |
| 7460 | 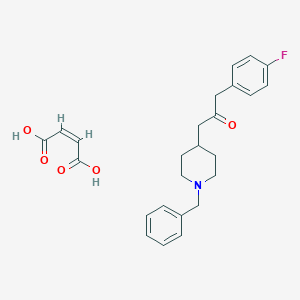 CHEMBL334466 CHEMBL334466 | C25H28FNO5 | 441.499 | 7 / 2 | N/A | N/A |
| 9740 | 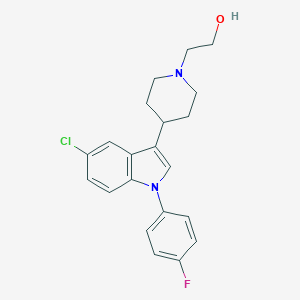 CHEMBL430151 CHEMBL430151 | C21H22ClFN2O | 372.868 | 3 / 1 | 4.3 | Yes |
| 459302 | 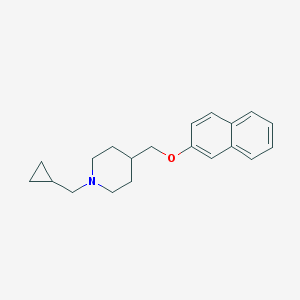 CHEMBL139578 CHEMBL139578 | C20H25NO | 295.426 | 2 / 0 | 4.7 | Yes |
| 10042 | 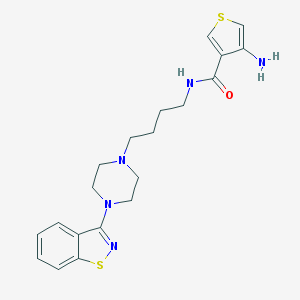 CHEMBL143075 CHEMBL143075 | C20H25N5OS2 | 415.574 | 7 / 2 | 3.8 | Yes |
| 10978 | 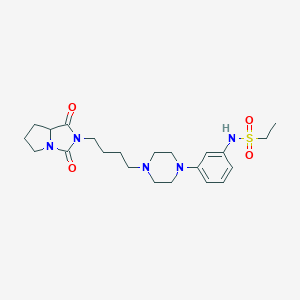 EF-7412 EF-7412 | C22H33N5O4S | 463.597 | 7 / 1 | 1.5 | Yes |
| 555522 | 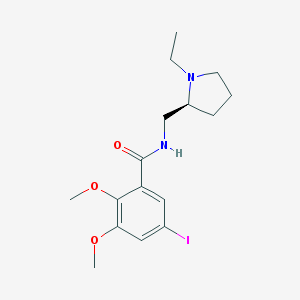 Epidepride Epidepride | C16H23IN2O3 | 418.275 | 4 / 1 | 2.1 | Yes |
| 11288 |  CHEMBL439609 CHEMBL439609 | C18H30N2O2 | 306.45 | 4 / 1 | N/A | N/A |
| 12313 |  CHEMBL13051 CHEMBL13051 | C28H34ClFN4O | 497.055 | 3 / 1 | 5.5 | No |
| 12938 |  CHEMBL267304 CHEMBL267304 | C25H29FN4O | 420.532 | 3 / 1 | 3.8 | Yes |
| 14820 |  CHEMBL138281 CHEMBL138281 | C21H21F4NO | 379.399 | 6 / 0 | 4.8 | Yes |
| 459364 | 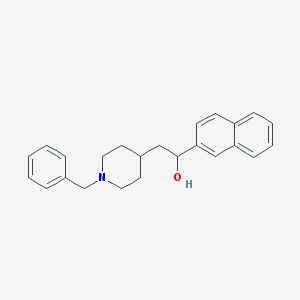 CHEMBL137250 CHEMBL137250 | C24H27NO | 345.486 | 2 / 1 | 5.0 | Yes |
| 20181 | 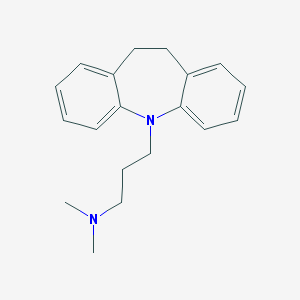 imipramine imipramine | C19H24N2 | 280.415 | 2 / 0 | 4.8 | Yes |
| 459401 | 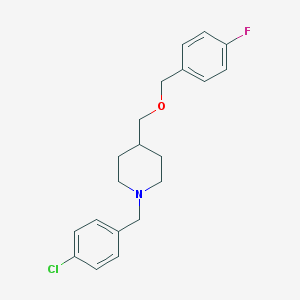 CHEMBL539567 CHEMBL539567 | C20H23ClFNO | 347.858 | 3 / 0 | 4.6 | Yes |
| 21653 | 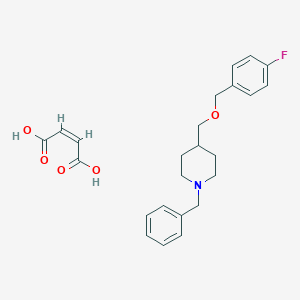 CHEMBL136495 CHEMBL136495 | C24H28FNO5 | 429.488 | 7 / 2 | N/A | N/A |
| 555567 | 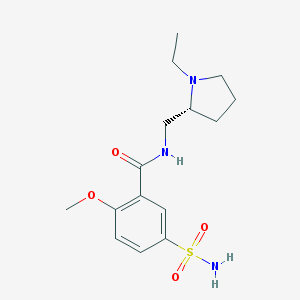 (+)-sulpiride (+)-sulpiride | C15H23N3O4S | 341.426 | 6 / 2 | 0.6 | Yes |
| 555568 | 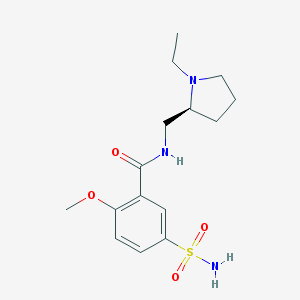 Levosulpiride Levosulpiride | C15H23N3O4S | 341.426 | 6 / 2 | 0.6 | Yes |
| 23365 | 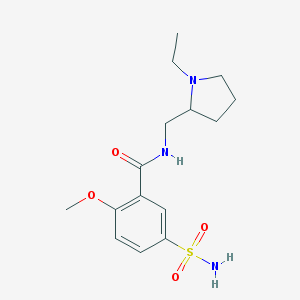 sulpiride sulpiride | C15H23N3O4S | 341.426 | 6 / 2 | 0.6 | Yes |
| 23455 | 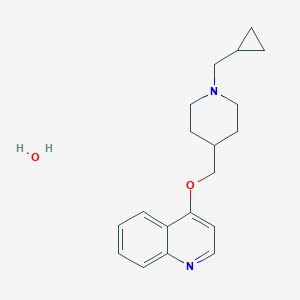 CHEMBL137502 CHEMBL137502 | C19H26N2O2 | 314.429 | 4 / 1 | N/A | N/A |
| 459438 | 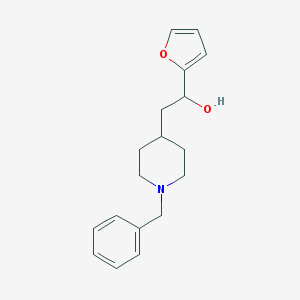 CHEMBL343926 CHEMBL343926 | C18H23NO2 | 285.387 | 3 / 1 | 2.8 | Yes |
| 26210 | 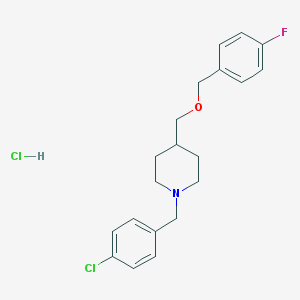 CHEMBL539567 CHEMBL539567 | C20H24Cl2FNO | 384.316 | 3 / 1 | N/A | N/A |
| 26768 | 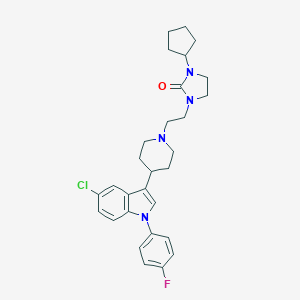 CHEMBL268971 CHEMBL268971 | C29H34ClFN4O | 509.066 | 3 / 0 | 5.5 | No |
| 27561 | 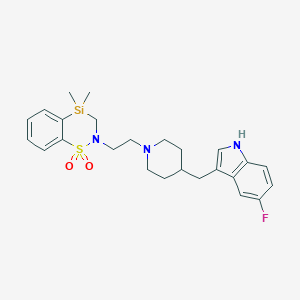 CHEMBL57514 CHEMBL57514 | C25H32FN3O2SSi | 485.693 | 5 / 1 | N/A | N/A |
| 459477 | 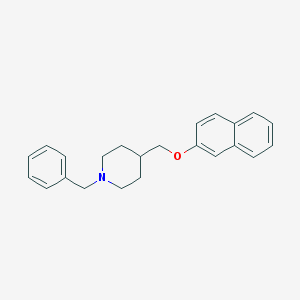 CHEMBL424281 CHEMBL424281 | C23H25NO | 331.459 | 2 / 0 | 5.4 | No |
| 555612 | 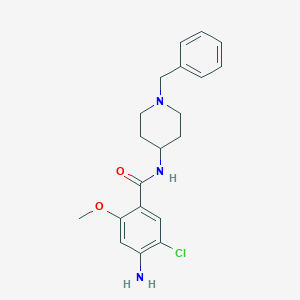 Clebopride Clebopride | C20H24ClN3O2 | 373.881 | 4 / 2 | 3.2 | Yes |
| 34704 | 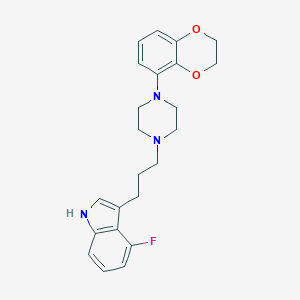 CHEMBL193854 CHEMBL193854 | C23H26FN3O2 | 395.478 | 5 / 1 | 4.1 | Yes |
| 459564 | 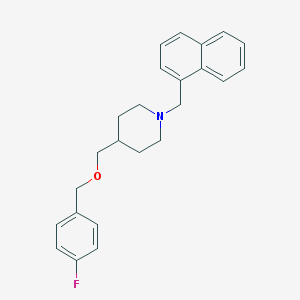 CHEMBL545552 CHEMBL545552 | C24H26FNO | 363.476 | 3 / 0 | 5.2 | No |
| 459568 |  CHEMBL136064 CHEMBL136064 | C23H25FN2O | 364.464 | 3 / 1 | 4.7 | Yes |
| 41534 |  CHEMBL543917 CHEMBL543917 | C18H25ClF3NO2 | 379.848 | 6 / 2 | N/A | N/A |
| 459595 |  138030-84-9 138030-84-9 | C20H25NO | 295.426 | 2 / 0 | 3.8 | Yes |
| 42547 |  CHEMBL136381 CHEMBL136381 | C18H23FN2O2 | 318.392 | 5 / 1 | N/A | N/A |
| 459648 |  CHEMBL543437 CHEMBL543437 | C21H24F3NO | 363.424 | 5 / 1 | 4.6 | Yes |
| 48385 |  CHEMBL136789 CHEMBL136789 | C21H25NOS | 339.497 | 3 / 0 | 4.3 | Yes |
| 459660 |  CHEMBL545549 CHEMBL545549 | C17H25NOS | 291.453 | 3 / 0 | 3.9 | Yes |
| 459662 |  CHEMBL554678 CHEMBL554678 | C23H31NO | 337.507 | 2 / 0 | 5.0 | Yes |
| 49441 |  CHEMBL136701 CHEMBL136701 | C22H30FNO2 | 359.485 | 4 / 2 | N/A | N/A |
| 459663 |  CHEMBL80656 CHEMBL80656 | C22H29NO | 323.48 | 2 / 0 | 4.7 | Yes |
| 49623 |  CHEMBL369984 CHEMBL369984 | C23H26FN3O2 | 395.478 | 5 / 1 | 4.1 | Yes |
| 459668 |  CHEMBL555975 CHEMBL555975 | C21H31NO | 313.485 | 2 / 0 | 4.7 | Yes |
| 459670 | 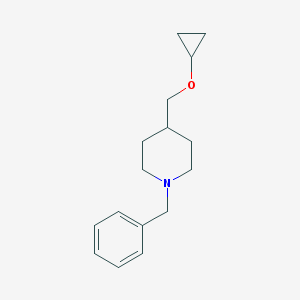 CHEMBL1184322 CHEMBL1184322 | C16H23NO | 245.366 | 2 / 0 | 2.9 | Yes |
| 51090 | 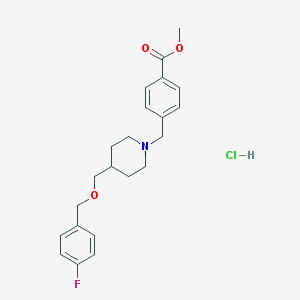 CHEMBL558819 CHEMBL558819 | C22H27ClFNO3 | 407.91 | 5 / 1 | N/A | N/A |
| 52266 | 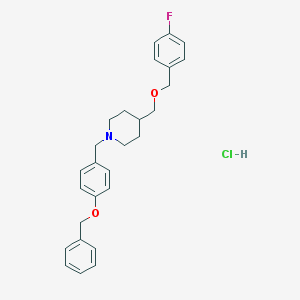 CHEMBL545081 CHEMBL545081 | C27H31ClFNO2 | 455.998 | 4 / 1 | N/A | N/A |
| 52635 | 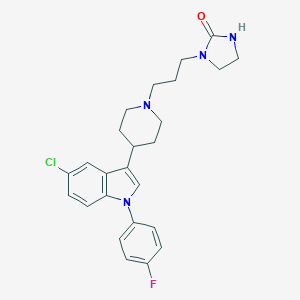 CHEMBL267549 CHEMBL267549 | C25H28ClFN4O | 454.974 | 3 / 1 | 4.4 | Yes |
| 55362 | 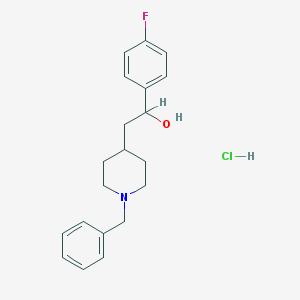 CHEMBL555443 CHEMBL555443 | C20H25ClFNO | 349.874 | 3 / 2 | N/A | N/A |
| 459745 | 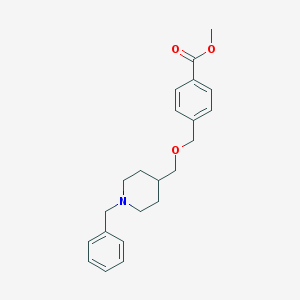 CHEMBL545781 CHEMBL545781 | C22H27NO3 | 353.462 | 4 / 0 | 3.7 | Yes |
| 57170 | 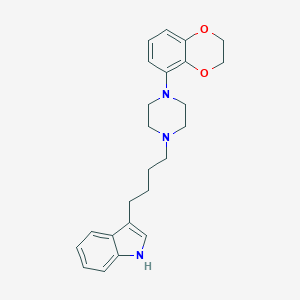 CHEMBL194844 CHEMBL194844 | C24H29N3O2 | 391.515 | 4 / 1 | 4.4 | Yes |
| 59770 | 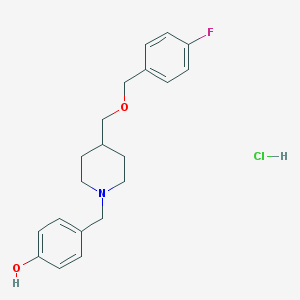 CHEMBL544147 CHEMBL544147 | C20H25ClFNO2 | 365.873 | 4 / 2 | N/A | N/A |
| 459772 | 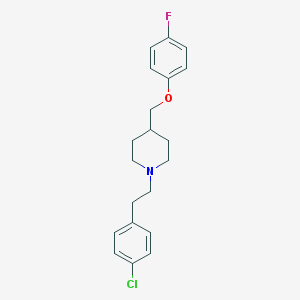 CHEMBL555865 CHEMBL555865 | C20H23ClFNO | 347.858 | 3 / 0 | 5.4 | No |
| 60991 | 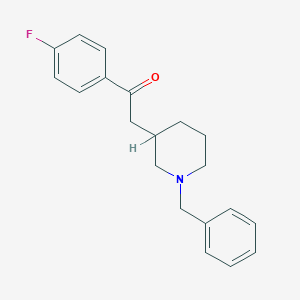 CHEMBL136781 CHEMBL136781 | C20H22FNO | 311.4 | 3 / 0 | 3.9 | Yes |
| 555714 | 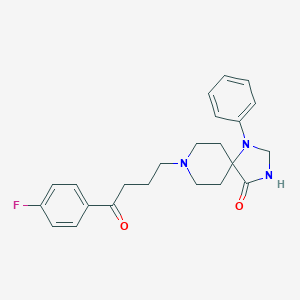 spiperone spiperone | C23H26FN3O2 | 395.478 | 5 / 1 | 3.0 | Yes |
| 459794 | 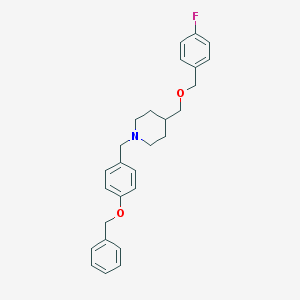 CHEMBL545081 CHEMBL545081 | C27H30FNO2 | 419.54 | 4 / 0 | 5.4 | No |
| 66460 | 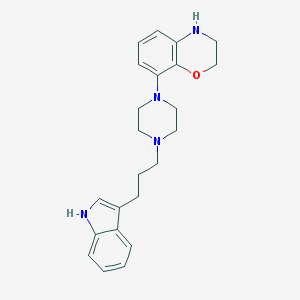 CHEMBL197665 CHEMBL197665 | C23H28N4O | 376.504 | 4 / 2 | 4.0 | Yes |
| 459830 | 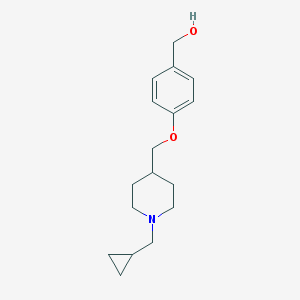 CHEMBL136877 CHEMBL136877 | C17H25NO2 | 275.392 | 3 / 1 | 2.5 | Yes |
| 459831 | 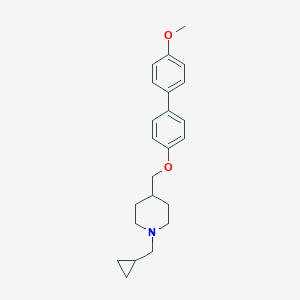 CHEMBL136205 CHEMBL136205 | C23H29NO2 | 351.49 | 3 / 0 | 5.0 | Yes |
| 70292 | 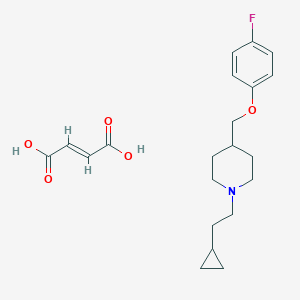 CHEMBL423353 CHEMBL423353 | C21H28FNO5 | 393.455 | 7 / 2 | N/A | N/A |
| 71696 | 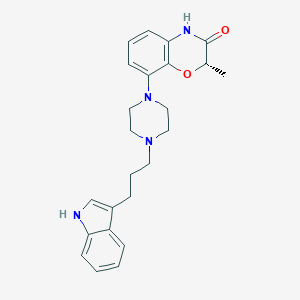 CHEMBL198224 CHEMBL198224 | C24H28N4O2 | 404.514 | 4 / 2 | 3.6 | Yes |
| 71697 | 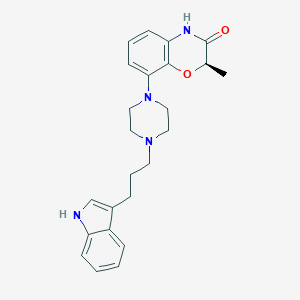 CHEMBL198216 CHEMBL198216 | C24H28N4O2 | 404.514 | 4 / 2 | 3.6 | Yes |
| 71699 | 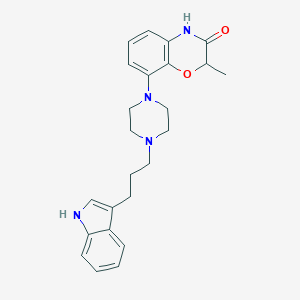 CHEMBL198070 CHEMBL198070 | C24H28N4O2 | 404.514 | 4 / 2 | 3.6 | Yes |
| 459908 | 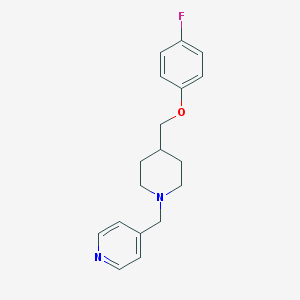 CHEMBL136381 CHEMBL136381 | C18H21FN2O | 300.377 | 4 / 0 | 3.2 | Yes |
| 459913 | 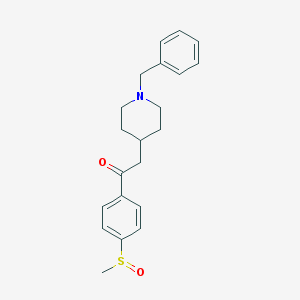 CHEMBL337148 CHEMBL337148 | C21H25NO2S | 355.496 | 4 / 0 | 2.9 | Yes |
| 78316 | 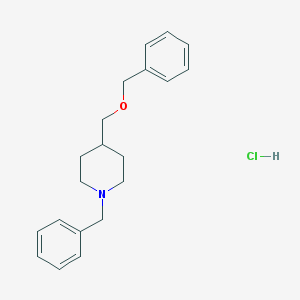 CHEMBL544146 CHEMBL544146 | C20H26ClNO | 331.884 | 2 / 1 | N/A | N/A |
| 78733 | 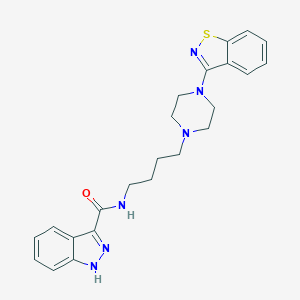 CHEMBL343838 CHEMBL343838 | C23H26N6OS | 434.562 | 6 / 2 | 4.2 | Yes |
| 78987 | 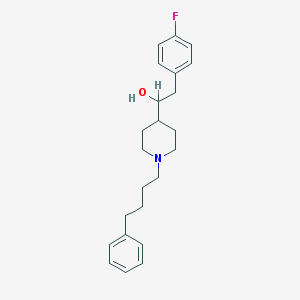 CHEMBL136622 CHEMBL136622 | C23H30FNO | 355.497 | 3 / 1 | 5.1 | No |
| 81087 | 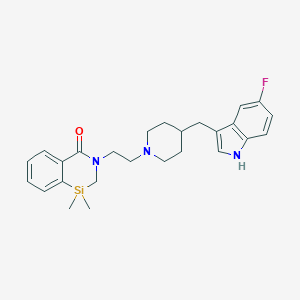 CHEMBL56571 CHEMBL56571 | C26H32FN3OSi | 449.645 | 3 / 1 | N/A | N/A |
| 459954 | 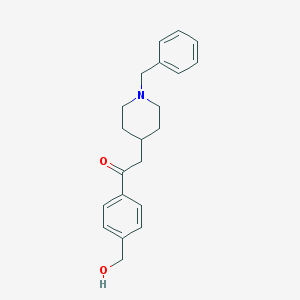 CHEMBL136388 CHEMBL136388 | C21H25NO2 | 323.436 | 3 / 1 | 2.9 | Yes |
| 83449 | 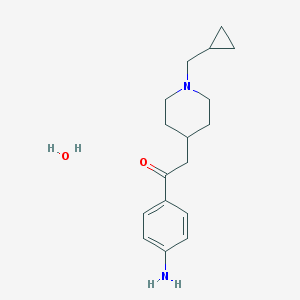 CHEMBL137163 CHEMBL137163 | C17H26N2O2 | 290.407 | 4 / 2 | N/A | N/A |
| 83589 | 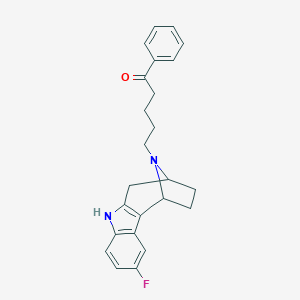 CHEMBL37385 CHEMBL37385 | C24H25FN2O | 376.475 | 3 / 1 | 4.7 | Yes |
| 555796 | 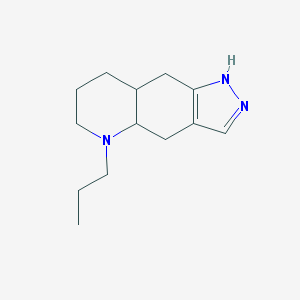 (A+/-)-Quinpirole dihydrochloride (A+/-)-Quinpirole dihydrochloride | C13H21N3 | 219.332 | 2 / 1 | 2.3 | Yes |
| 555799 | 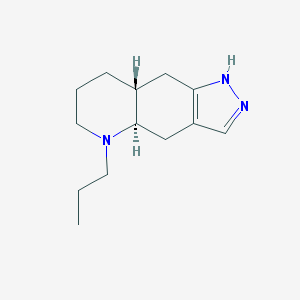 QUINPIROLE QUINPIROLE | C13H21N3 | 219.332 | 2 / 1 | 2.3 | Yes |
| 88698 | 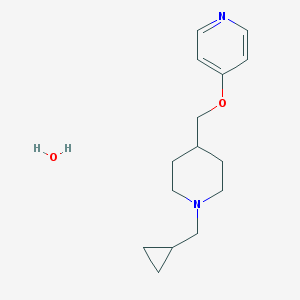 CHEMBL139102 CHEMBL139102 | C15H24N2O2 | 264.369 | 4 / 1 | N/A | N/A |
| 89291 | 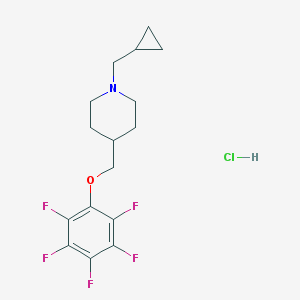 CHEMBL543438 CHEMBL543438 | C16H19ClF5NO | 371.776 | 7 / 1 | N/A | N/A |
| 89752 | 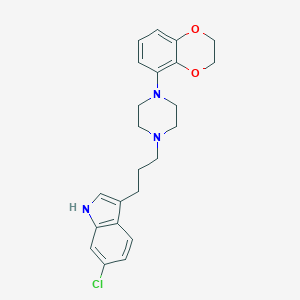 CHEMBL194240 CHEMBL194240 | C23H26ClN3O2 | 411.93 | 4 / 1 | 4.6 | Yes |
| 460026 | 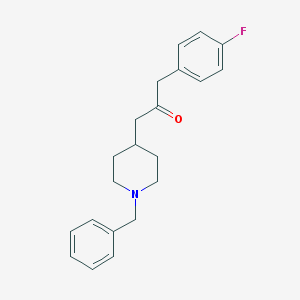 CHEMBL334466 CHEMBL334466 | C21H24FNO | 325.427 | 3 / 0 | 3.9 | Yes |
| 91942 | 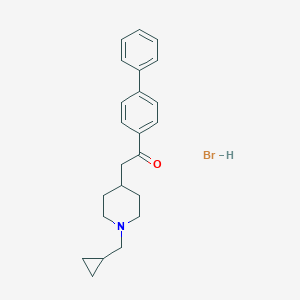 CHEMBL544372 CHEMBL544372 | C23H28BrNO | 414.387 | 2 / 1 | N/A | N/A |
| 460040 | 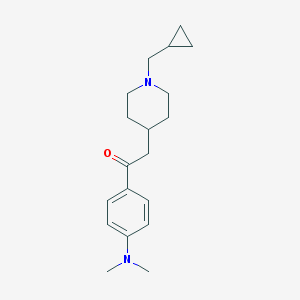 CHEMBL543206 CHEMBL543206 | C19H28N2O | 300.446 | 3 / 0 | 3.2 | Yes |
| 92893 | 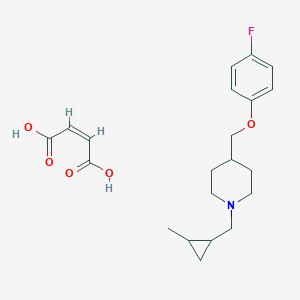 CHEMBL137085 CHEMBL137085 | C21H28FNO5 | 393.455 | 7 / 2 | N/A | N/A |
| 94368 | 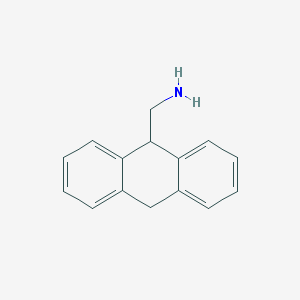 CHEMBL47482 CHEMBL47482 | C15H15N | 209.292 | 1 / 1 | 2.7 | Yes |
| 95717 | 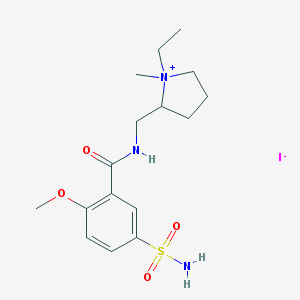 CHEMBL170310 CHEMBL170310 | C16H26IN3O4S | 483.365 | 6 / 2 | N/A | N/A |
| 460069 | 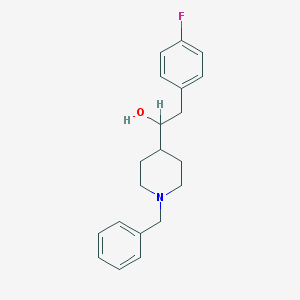 CHEMBL545546 CHEMBL545546 | C20H24FNO | 313.416 | 3 / 1 | 3.9 | Yes |
| 99822 | 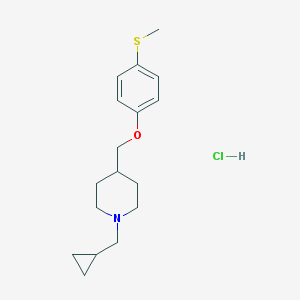 CHEMBL545549 CHEMBL545549 | C17H26ClNOS | 327.911 | 3 / 1 | N/A | N/A |
| 101529 | 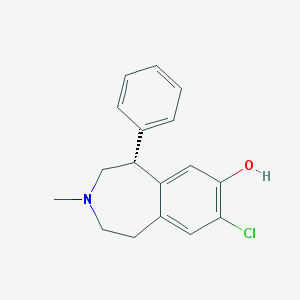 UNII-UGT5535REQ UNII-UGT5535REQ | C17H18ClNO | 287.787 | 2 / 1 | 4.0 | Yes |
| 102657 | 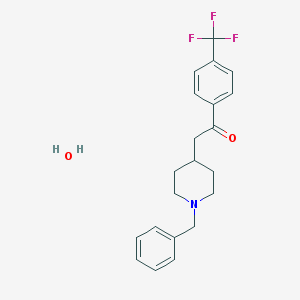 CHEMBL136068 CHEMBL136068 | C21H24F3NO2 | 379.423 | 6 / 1 | N/A | N/A |
| 104260 | 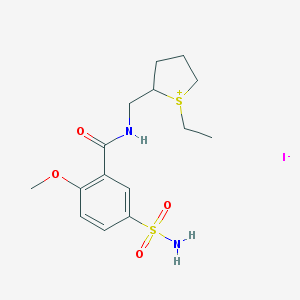 CHEMBL170622 CHEMBL170622 | C15H23IN2O4S2 | 486.383 | 6 / 2 | N/A | N/A |
| 104621 | 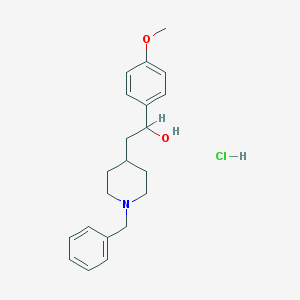 CHEMBL539042 CHEMBL539042 | C21H28ClNO2 | 361.91 | 3 / 2 | N/A | N/A |
| 104928 | 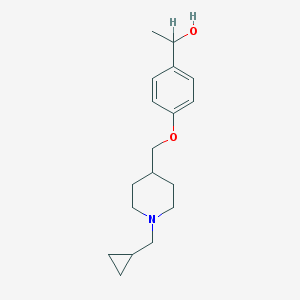 CHEMBL136702 CHEMBL136702 | C18H27NO2 | 289.419 | 3 / 1 | 2.9 | Yes |
| 105463 | 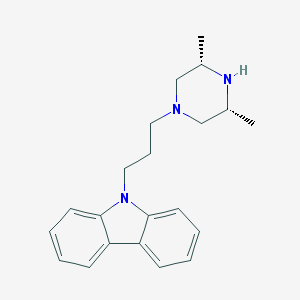 Rimcazole Rimcazole | C21H27N3 | 321.468 | 2 / 1 | 3.9 | Yes |
| 107695 | 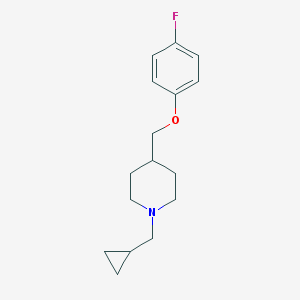 CHEMBL335327 CHEMBL335327 | C16H22FNO | 263.356 | 3 / 0 | 3.5 | Yes |
| 108110 |  CHEMBL423355 CHEMBL423355 | C18H24N2O | 284.403 | 2 / 1 | 3.6 | Yes |
| 108218 |  CHEMBL383396 CHEMBL383396 | C24H28N4O | 388.515 | 3 / 2 | 3.4 | Yes |
| 109133 |  SERTINDOLE SERTINDOLE | C24H26ClFN4O | 440.947 | 3 / 1 | 4.1 | Yes |
| 111136 |  CHEMBL300786 CHEMBL300786 | C24H26FN3O | 391.49 | 3 / 1 | 3.9 | Yes |
| 112065 | 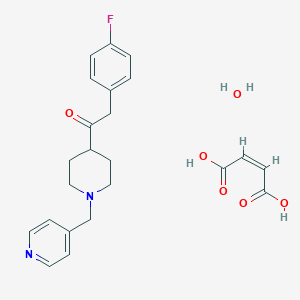 CHEMBL337124 CHEMBL337124 | C23H27FN2O6 | 446.475 | 9 / 3 | N/A | N/A |
| 113580 | 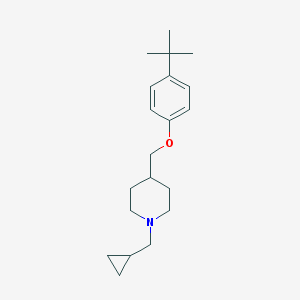 CHEMBL137563 CHEMBL137563 | C20H31NO | 301.474 | 2 / 0 | 5.1 | No |
| 115624 | 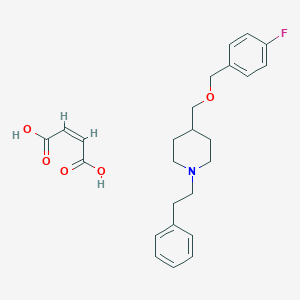 CHEMBL136963 CHEMBL136963 | C25H30FNO5 | 443.515 | 7 / 2 | N/A | N/A |
| 117900 | 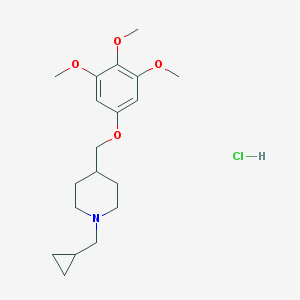 CHEMBL541603 CHEMBL541603 | C19H30ClNO4 | 371.902 | 5 / 1 | N/A | N/A |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417