

You can:
| Name | Metabotropic glutamate receptor 4 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | GRM4 |
| Synonym | mGluR4 mGlu4 receptor GPRC1D glutamate receptor |
| Disease | Anxiety disorder Neurological disease |
| Length | 912 |
| Amino acid sequence | MPGKRGLGWWWARLPLCLLLSLYGPWMPSSLGKPKGHPHMNSIRIDGDITLGGLFPVHGRGSEGKPCGELKKEKGIHRLEAMLFALDRINNDPDLLPNITLGARILDTCSRDTHALEQSLTFVQALIEKDGTEVRCGSGGPPIITKPERVVGVIGASGSSVSIMVANILRLFKIPQISYASTAPDLSDNSRYDFFSRVVPSDTYQAQAMVDIVRALKWNYVSTVASEGSYGESGVEAFIQKSREDGGVCIAQSVKIPREPKAGEFDKIIRRLLETSNARAVIIFANEDDIRRVLEAARRANQTGHFFWMGSDSWGSKIAPVLHLEEVAEGAVTILPKRMSVRGFDRYFSSRTLDNNRRNIWFAEFWEDNFHCKLSRHALKKGSHVKKCTNRERIGQDSAYEQEGKVQFVIDAVYAMGHALHAMHRDLCPGRVGLCPRMDPVDGTQLLKYIRNVNFSGIAGNPVTFNENGDAPGRYDIYQYQLRNDSAEYKVIGSWTDHLHLRIERMHWPGSGQQLPRSICSLPCQPGERKKTVKGMPCCWHCEPCTGYQYQVDRYTCKTCPYDMRPTENRTGCRPIPIIKLEWGSPWAVLPLFLAVVGIAATLFVVITFVRYNDTPIVKASGRELSYVLLAGIFLCYATTFLMIAEPDLGTCSLRRIFLGLGMSISYAALLTKTNRIYRIFEQGKRSVSAPRFISPASQLAITFSLISLQLLGICVWFVVDPSHSVVDFQDQRTLDPRFARGVLKCDISDLSLICLLGYSMLLMVTCTVYAIKTRGVPETFNEAKPIGFTMYTTCIVWLAFIPIFFGTSQSADKLYIQTTTLTVSVSLSASVSLGMLYMPKVYIILFHPEQNVPKRKRSLKAVVTAATMSNKFTQKGNFRPNGEAKSELCENLEAPALATKQTYVTYTNHAI |
| UniProt | Q14833 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | T99402 |
| ChEMBL | CHEMBL2736 |
| IUPHAR | 292 |
| DrugBank | BE0000833 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 441 | 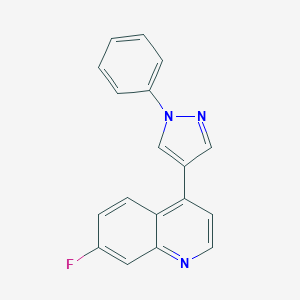 CHEMBL2023454 CHEMBL2023454 | C18H12FN3 | 289.313 | 3 / 0 | 3.9 | Yes |
| 517322 | 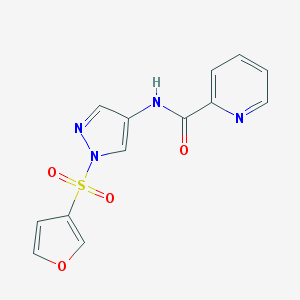 CHEMBL3952345 CHEMBL3952345 | C13H10N4O4S | 318.307 | 6 / 1 | 0.6 | Yes |
| 517323 | 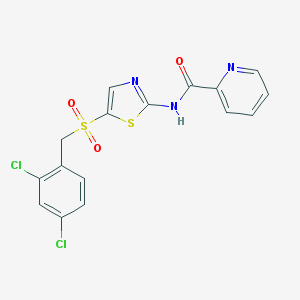 CHEMBL3939625 CHEMBL3939625 | C16H11Cl2N3O3S2 | 428.302 | 6 / 1 | 3.7 | Yes |
| 463129 | 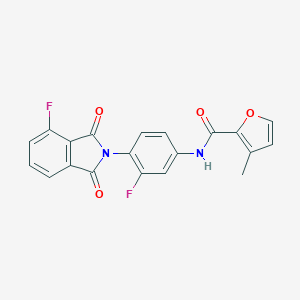 CHEMBL3634421 CHEMBL3634421 | C20H12F2N2O4 | 382.323 | 6 / 1 | 3.3 | Yes |
| 555483 | 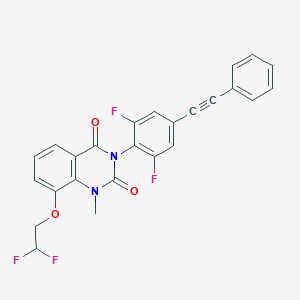 SCHEMBL16607448 SCHEMBL16607448 | C25H16F4N2O3 | 468.408 | 7 / 0 | 5.4 | No |
| 459247 | 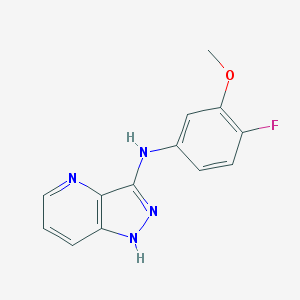 SCHEMBL2360200 SCHEMBL2360200 | C13H11FN4O | 258.256 | 5 / 2 | 2.5 | Yes |
| 2374 | 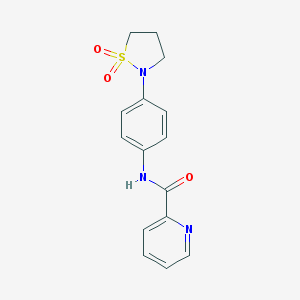 CHEMBL1921958 CHEMBL1921958 | C15H15N3O3S | 317.363 | 5 / 1 | 1.3 | Yes |
| 3883 | 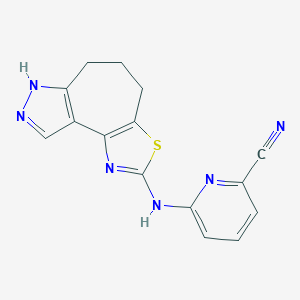 CHEMBL1830907 CHEMBL1830907 | C15H12N6S | 308.363 | 6 / 2 | 2.8 | Yes |
| 4943 | 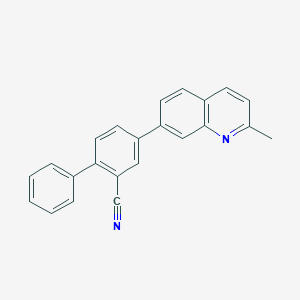 CHEMBL1099113 CHEMBL1099113 | C23H16N2 | 320.395 | 2 / 0 | 5.6 | No |
| 5420 | 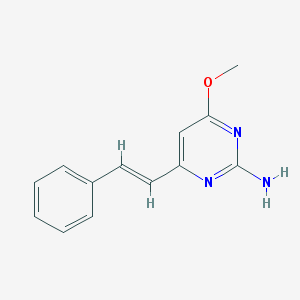 CHEMBL1209518 CHEMBL1209518 | C13H13N3O | 227.267 | 4 / 1 | 2.5 | Yes |
| 5717 | 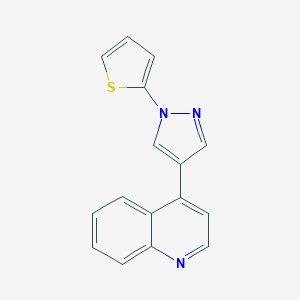 CHEMBL2022873 CHEMBL2022873 | C16H11N3S | 277.345 | 3 / 0 | 3.8 | Yes |
| 6506 | 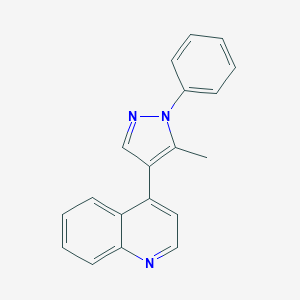 CHEMBL2023452 CHEMBL2023452 | C19H15N3 | 285.35 | 2 / 0 | 4.2 | Yes |
| 441958 | 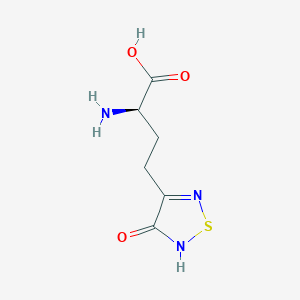 CHEMBL334842 CHEMBL334842 | C6H9N3O3S | 203.216 | 6 / 3 | -3.4 | Yes |
| 441963 | 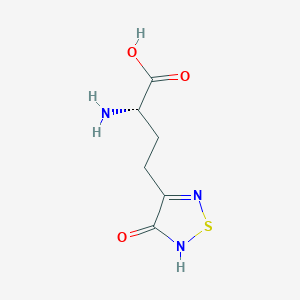 CHEMBL131922 CHEMBL131922 | C6H9N3O3S | 203.216 | 6 / 3 | -3.4 | Yes |
| 521649 | 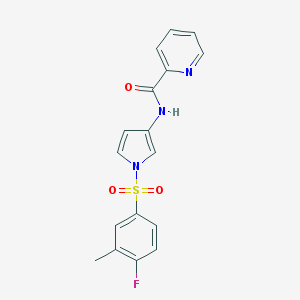 CHEMBL3809272 CHEMBL3809272 | C17H14FN3O3S | 359.375 | 5 / 1 | 2.5 | Yes |
| 536125 | 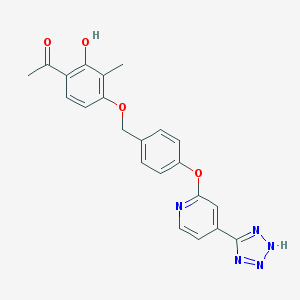 CHEMBL3899832 CHEMBL3899832 | C22H19N5O4 | 417.425 | 8 / 2 | 3.6 | Yes |
| 7085 | 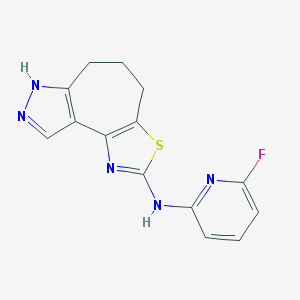 CHEMBL1830903 CHEMBL1830903 | C14H12FN5S | 301.343 | 6 / 2 | 3.2 | Yes |
| 459290 | 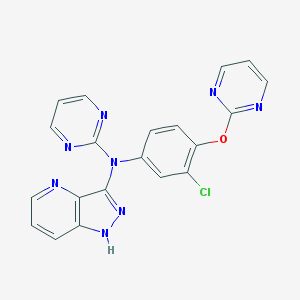 SCHEMBL6923581 SCHEMBL6923581 | C20H13ClN8O | 416.829 | 8 / 1 | 3.5 | Yes |
| 8266 | 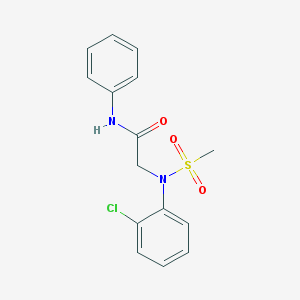 CHEMBL2208408 CHEMBL2208408 | C15H15ClN2O3S | 338.806 | 4 / 1 | 2.6 | Yes |
| 8296 | 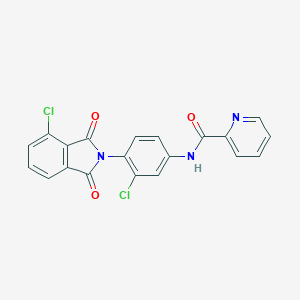 VU0405623-1 VU0405623-1 | C20H11Cl2N3O3 | 412.226 | 4 / 1 | 3.8 | Yes |
| 8729 | 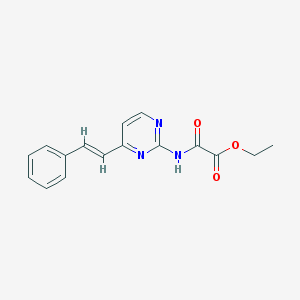 CHEMBL1209746 CHEMBL1209746 | C16H15N3O3 | 297.314 | 5 / 1 | 2.7 | Yes |
| 517364 | 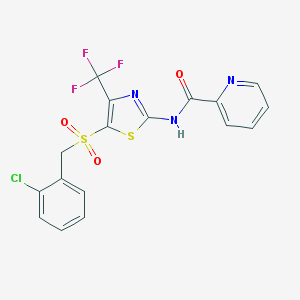 CHEMBL3986822 CHEMBL3986822 | C17H11ClF3N3O3S2 | 461.858 | 9 / 1 | 4.0 | Yes |
| 442079 | 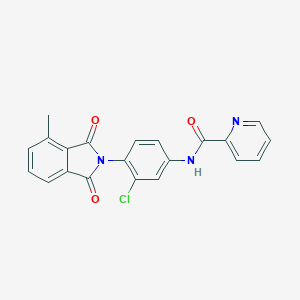 CHEMBL3357575 CHEMBL3357575 | C21H14ClN3O3 | 391.811 | 4 / 1 | 3.5 | Yes |
| 9585 | 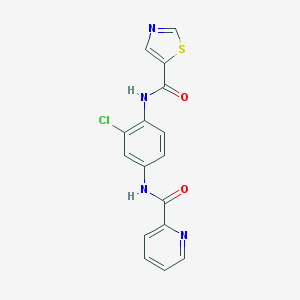 CHEMBL1909433 CHEMBL1909433 | C16H11ClN4O2S | 358.8 | 5 / 2 | 2.8 | Yes |
| 10076 | 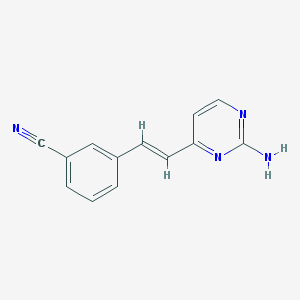 CHEMBL1209663 CHEMBL1209663 | C13H10N4 | 222.251 | 4 / 1 | 1.9 | Yes |
| 10131 | 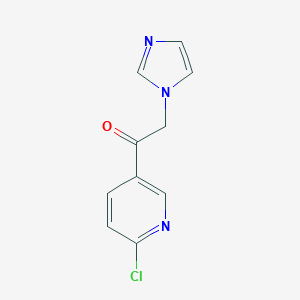 1-(6-chloro-3-pyridyl)-2-imidazol-1-ylethanone 1-(6-chloro-3-pyridyl)-2-imidazol-1-ylethanone | C10H8ClN3O | 221.644 | 3 / 0 | 1.4 | Yes |
| 10197 | 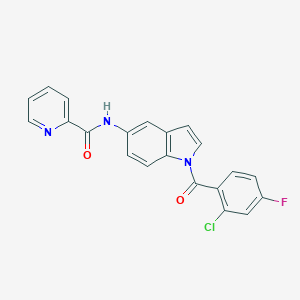 CHEMBL1672256 CHEMBL1672256 | C21H13ClFN3O2 | 393.802 | 4 / 1 | 4.4 | Yes |
| 464129 | 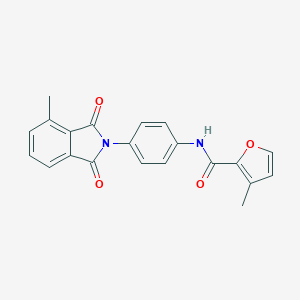 CHEMBL3634415 CHEMBL3634415 | C21H16N2O4 | 360.369 | 4 / 1 | 3.4 | Yes |
| 10405 | 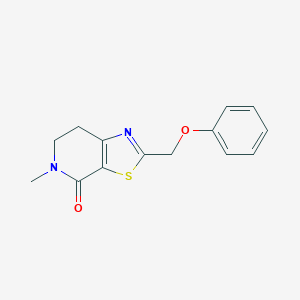 CHEMBL2426616 CHEMBL2426616 | C14H14N2O2S | 274.338 | 4 / 0 | 2.3 | Yes |
| 10846 | 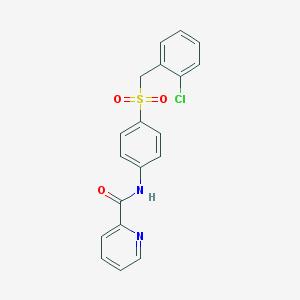 CHEMBL1223384 CHEMBL1223384 | C19H15ClN2O3S | 386.85 | 4 / 1 | 3.4 | Yes |
| 459321 | 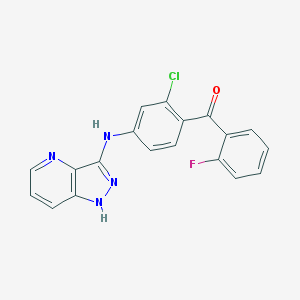 SCHEMBL6923296 SCHEMBL6923296 | C19H12ClFN4O | 366.78 | 5 / 2 | 4.5 | Yes |
| 459333 | 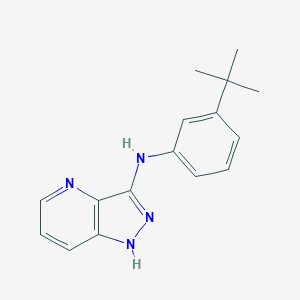 SCHEMBL6922895 SCHEMBL6922895 | C16H18N4 | 266.348 | 3 / 2 | 4.1 | Yes |
| 12904 | 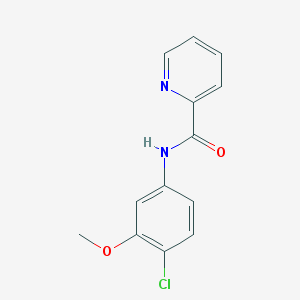 VU 0361737 VU 0361737 | C13H11ClN2O2 | 262.693 | 3 / 1 | 2.6 | Yes |
| 13246 | 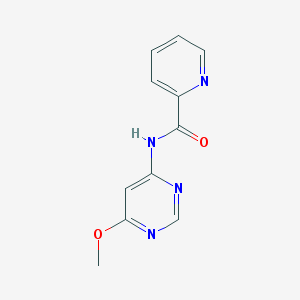 N-(6-methoxypyrimidin-4-yl)picolinamide N-(6-methoxypyrimidin-4-yl)picolinamide | C11H10N4O2 | 230.227 | 5 / 1 | 0.9 | Yes |
| 13317 | 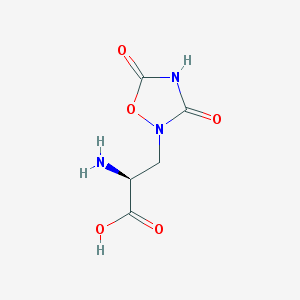 QUISQUALIC ACID QUISQUALIC ACID | C5H7N3O5 | 189.127 | 6 / 3 | -3.9 | Yes |
| 517389 | 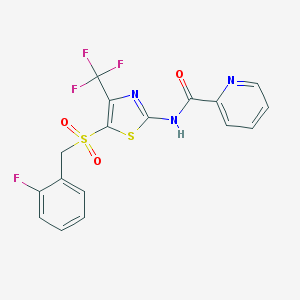 CHEMBL3958116 CHEMBL3958116 | C17H11F4N3O3S2 | 445.407 | 10 / 1 | 3.5 | Yes |
| 14004 | 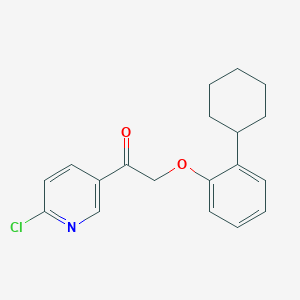 CHEMBL1922764 CHEMBL1922764 | C19H20ClNO2 | 329.824 | 3 / 0 | 5.7 | No |
| 517395 | 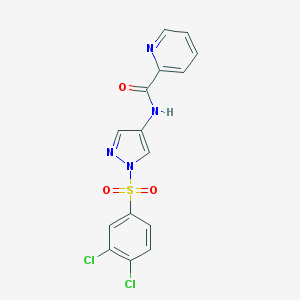 SCHEMBL1653189 SCHEMBL1653189 | C15H10Cl2N4O3S | 397.23 | 5 / 1 | 2.8 | Yes |
| 459348 | 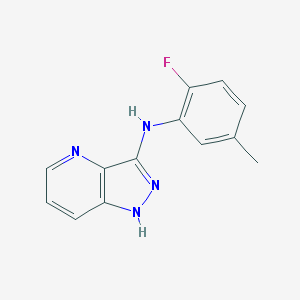 CHEMBL3930906 CHEMBL3930906 | C13H11FN4 | 242.257 | 4 / 2 | 2.9 | Yes |
| 517401 | 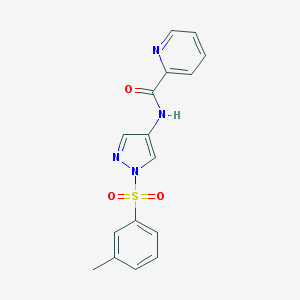 CHEMBL3974101 CHEMBL3974101 | C16H14N4O3S | 342.373 | 5 / 1 | 1.9 | Yes |
| 16007 | 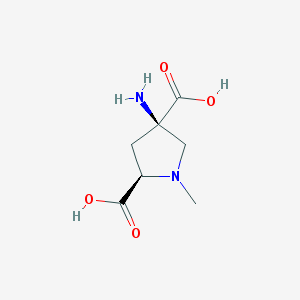 CHEMBL300330 CHEMBL300330 | C7H12N2O4 | 188.183 | 6 / 3 | -6.0 | Yes |
| 17083 | 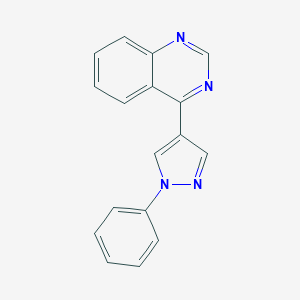 CHEMBL2023628 CHEMBL2023628 | C17H12N4 | 272.311 | 3 / 0 | 3.2 | Yes |
| 459375 | 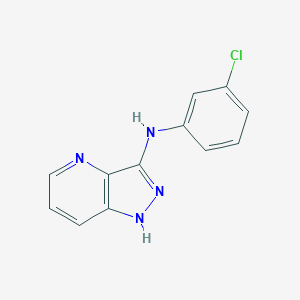 N-(3-chlorophenyl)-1H-Pyrazolo[4,3-b]pyridin-3-amine N-(3-chlorophenyl)-1H-Pyrazolo[4,3-b]pyridin-3-amine | C12H9ClN4 | 244.682 | 3 / 2 | 3.1 | Yes |
| 459376 | 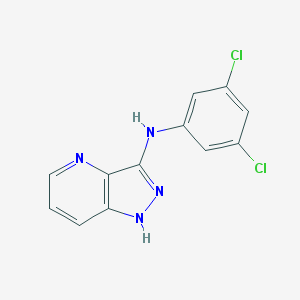 SCHEMBL6922917 SCHEMBL6922917 | C12H8Cl2N4 | 279.124 | 3 / 2 | 3.7 | Yes |
| 19020 | 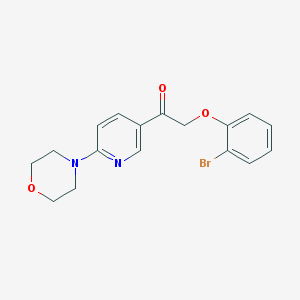 CHEMBL1922739 CHEMBL1922739 | C17H17BrN2O3 | 377.238 | 5 / 0 | 3.0 | Yes |
| 19296 | 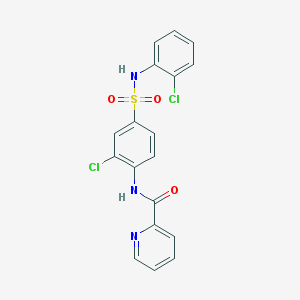 CHEMBL1223382 CHEMBL1223382 | C18H13Cl2N3O3S | 422.28 | 5 / 2 | 3.8 | Yes |
| 459386 | 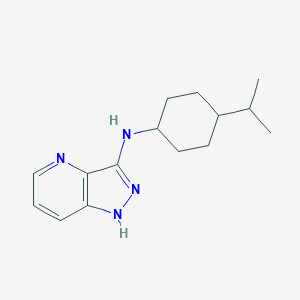 SCHEMBL6923592 SCHEMBL6923592 | C15H22N4 | 258.369 | 3 / 2 | 3.7 | Yes |
| 555557 | 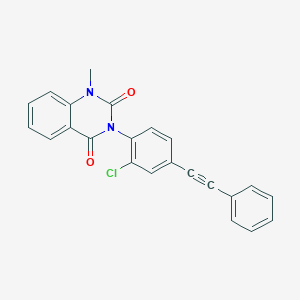 SCHEMBL16607464 SCHEMBL16607464 | C23H15ClN2O2 | 386.835 | 2 / 0 | 5.0 | Yes |
| 517427 | 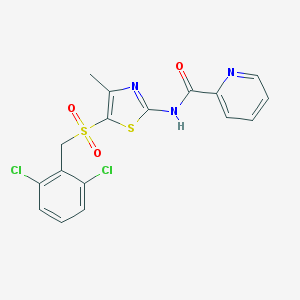 CHEMBL3984173 CHEMBL3984173 | C17H13Cl2N3O3S2 | 442.329 | 6 / 1 | 4.1 | Yes |
| 20310 | 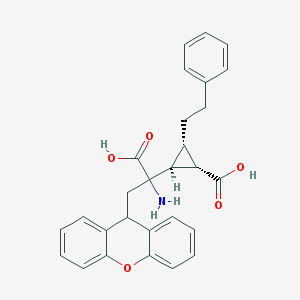 CHEMBL319279 CHEMBL319279 | C28H27NO5 | 457.526 | 6 / 3 | 2.1 | Yes |
| 536488 | 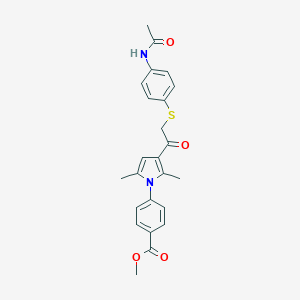 AC1N2AWG AC1N2AWG | C24H24N2O4S | 436.526 | 5 / 1 | 4.1 | Yes |
| 517436 | 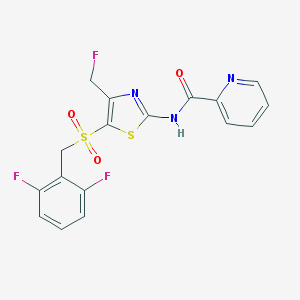 CHEMBL3956107 CHEMBL3956107 | C17H12F3N3O3S2 | 427.416 | 9 / 1 | 2.8 | Yes |
| 459407 | 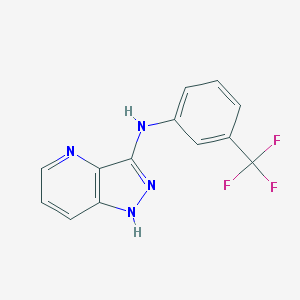 SCHEMBL6921970 SCHEMBL6921970 | C13H9F3N4 | 278.238 | 6 / 2 | 3.3 | Yes |
| 22395 | 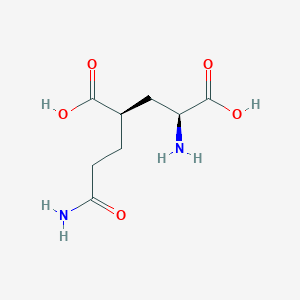 CHEMBL479804 CHEMBL479804 | C8H14N2O5 | 218.209 | 6 / 4 | -3.3 | Yes |
| 517446 | 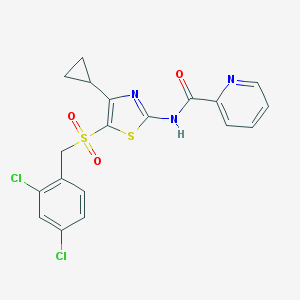 CHEMBL3960293 CHEMBL3960293 | C19H15Cl2N3O3S2 | 468.367 | 6 / 1 | 4.3 | Yes |
| 22860 | 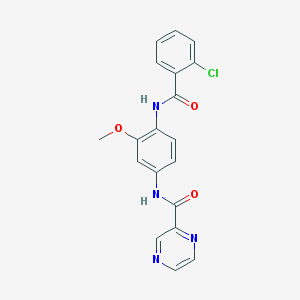 CHEMBL1672235 CHEMBL1672235 | C19H15ClN4O3 | 382.804 | 5 / 2 | 2.4 | Yes |
| 553358 | 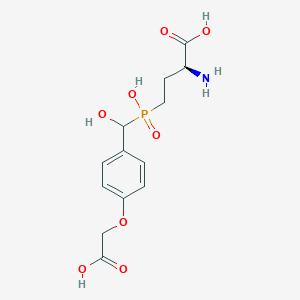 LSP4-2022 LSP4-2022 | C13H18NO8P | 347.26 | 9 / 5 | -3.8 | Yes |
| 25298 | 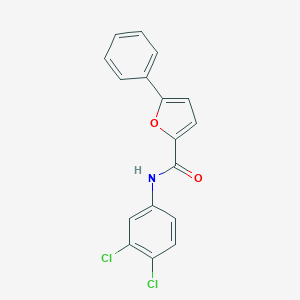 CHEMBL549670 CHEMBL549670 | C17H11Cl2NO2 | 332.18 | 2 / 1 | 5.1 | No |
| 522330 | 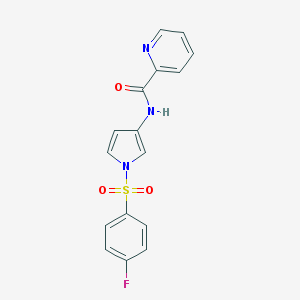 CHEMBL3809928 CHEMBL3809928 | C16H12FN3O3S | 345.348 | 5 / 1 | 2.1 | Yes |
| 459446 | 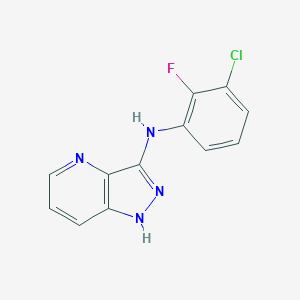 SCHEMBL2361692 SCHEMBL2361692 | C12H8ClFN4 | 262.672 | 4 / 2 | 3.2 | Yes |
| 27264 | 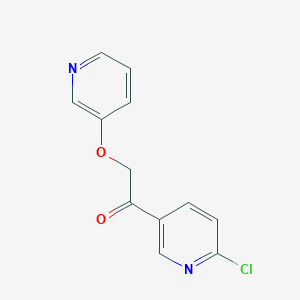 CHEMBL1922757 CHEMBL1922757 | C12H9ClN2O2 | 248.666 | 4 / 0 | 2.1 | Yes |
| 27577 | 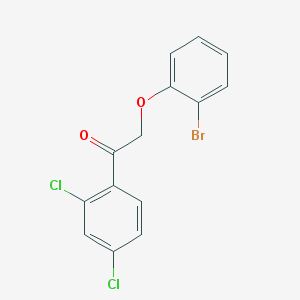 CHEMBL1922733 CHEMBL1922733 | C14H9BrCl2O2 | 360.028 | 2 / 0 | 5.3 | No |
| 522393 | 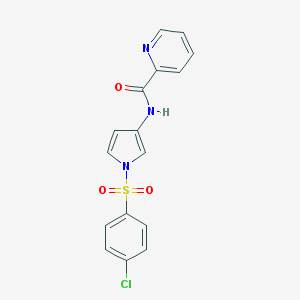 CHEMBL3808806 CHEMBL3808806 | C16H12ClN3O3S | 361.8 | 4 / 1 | 2.6 | Yes |
| 459465 | 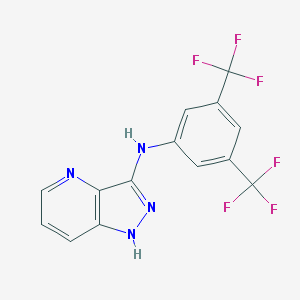 SCHEMBL6922222 SCHEMBL6922222 | C14H8F6N4 | 346.236 | 9 / 2 | 4.2 | Yes |
| 517471 | 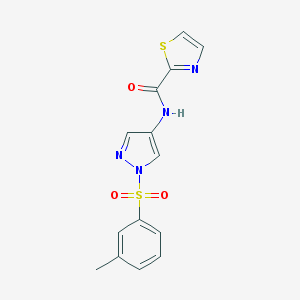 SCHEMBL1653827 SCHEMBL1653827 | C14H12N4O3S2 | 348.395 | 6 / 1 | 2.0 | Yes |
| 522459 | 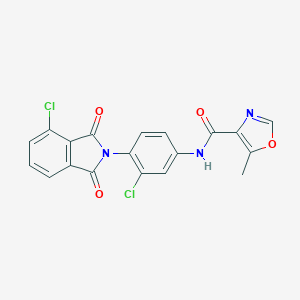 CHEMBL3759867 CHEMBL3759867 | C19H11Cl2N3O4 | 416.214 | 5 / 1 | 3.7 | Yes |
| 522475 | 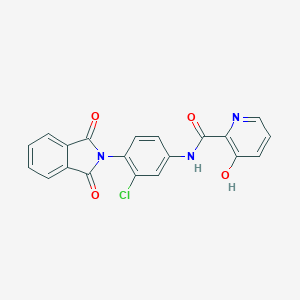 CHEMBL3800579 CHEMBL3800579 | C20H12ClN3O4 | 393.783 | 5 / 2 | 3.4 | Yes |
| 536769 | 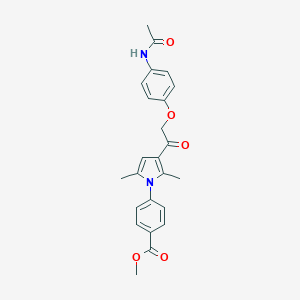 CHEMBL3560146 CHEMBL3560146 | C24H24N2O5 | 420.465 | 5 / 1 | 3.5 | Yes |
| 466733 |  CHEMBL3628111 CHEMBL3628111 | C20H12Cl2N2O4 | 415.226 | 4 / 1 | 4.3 | Yes |
| 459491 |  SCHEMBL6922868 SCHEMBL6922868 | C11H7ClFN5 | 263.66 | 5 / 2 | 2.4 | Yes |
| 522506 |  CHEMBL3800490 CHEMBL3800490 | C20H14Cl2N4O2 | 413.258 | 4 / 2 | 3.7 | Yes |
| 517486 |  CHEMBL3972975 CHEMBL3972975 | C18H14F3N3O3S2 | 441.443 | 9 / 1 | 3.7 | Yes |
| 517487 | 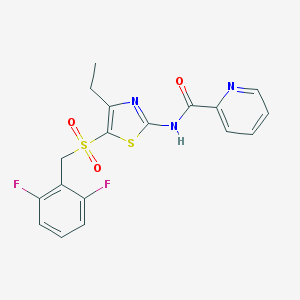 CHEMBL3808861 CHEMBL3808861 | C18H15F2N3O3S2 | 423.453 | 8 / 1 | 3.5 | Yes |
| 32490 | 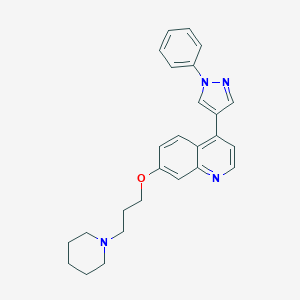 CHEMBL2023465 CHEMBL2023465 | C26H28N4O | 412.537 | 4 / 0 | 5.0 | Yes |
| 459512 | 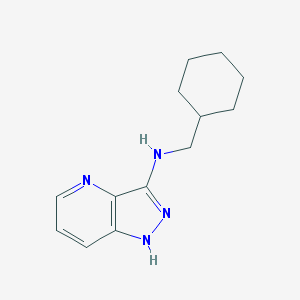 SCHEMBL6923917 SCHEMBL6923917 | C13H18N4 | 230.315 | 3 / 2 | 3.3 | Yes |
| 467004 | 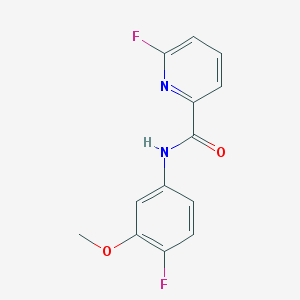 CHEMBL3609735 CHEMBL3609735 | C13H10F2N2O2 | 264.232 | 5 / 1 | 2.5 | Yes |
| 34970 | 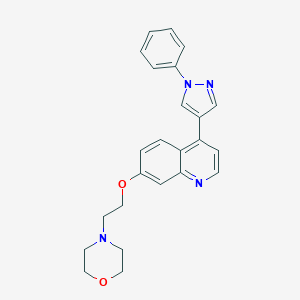 CHEMBL2023470 CHEMBL2023470 | C24H24N4O2 | 400.482 | 5 / 0 | 3.4 | Yes |
| 34972 | 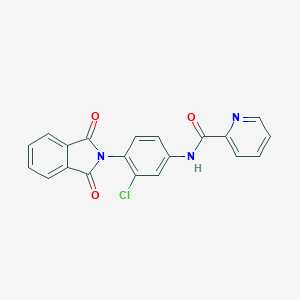 VU0405622-1 VU0405622-1 | C20H12ClN3O3 | 377.784 | 4 / 1 | 3.2 | Yes |
| 35338 | 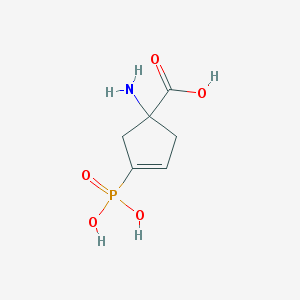 CHEMBL32972 CHEMBL32972 | C6H10NO5P | 207.122 | 6 / 4 | -4.6 | Yes |
| 35427 | 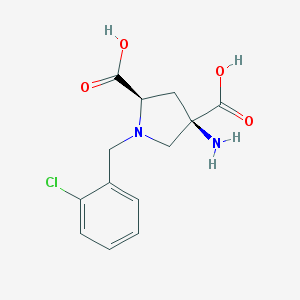 CHEMBL418462 CHEMBL418462 | C13H15ClN2O4 | 298.723 | 6 / 3 | -3.9 | Yes |
| 35747 | 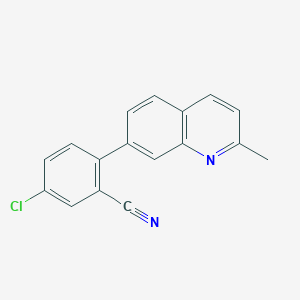 CHEMBL1099112 CHEMBL1099112 | C17H11ClN2 | 278.739 | 2 / 0 | 4.6 | Yes |
| 35785 | 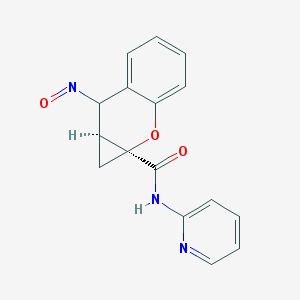 BDBM50388941 BDBM50388941 | C16H13N3O3 | 295.298 | 5 / 1 | 1.4 | Yes |
| 555615 | 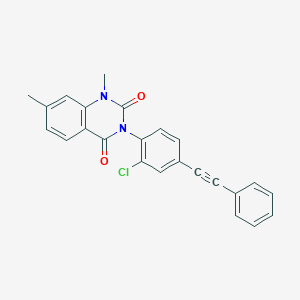 SCHEMBL16599137 SCHEMBL16599137 | C24H17ClN2O2 | 400.862 | 2 / 0 | 5.4 | No |
| 517500 | 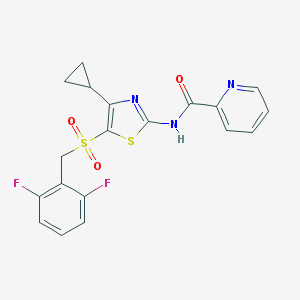 CHEMBL3904734 CHEMBL3904734 | C19H15F2N3O3S2 | 435.464 | 8 / 1 | 3.2 | Yes |
| 459543 | 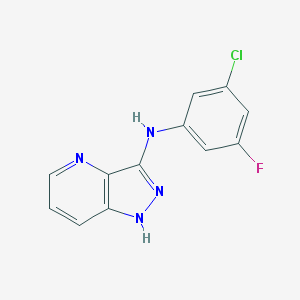 SCHEMBL6922218 SCHEMBL6922218 | C12H8ClFN4 | 262.672 | 4 / 2 | 3.2 | Yes |
| 517503 | 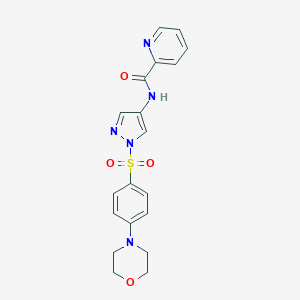 SCHEMBL1652013 SCHEMBL1652013 | C19H19N5O4S | 413.452 | 7 / 1 | 1.3 | Yes |
| 36682 | 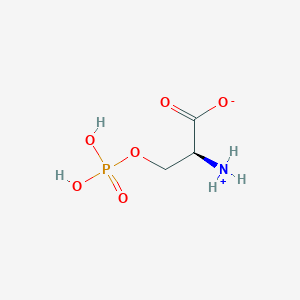 CID 57689797 CID 57689797 | C3H8NO6P | 185.072 | 6 / 3 | -4.5 | Yes |
| 467491 | 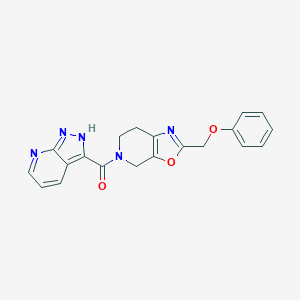 CHEMBL3605296 CHEMBL3605296 | C20H17N5O3 | 375.388 | 6 / 1 | 2.1 | Yes |
| 555616 | 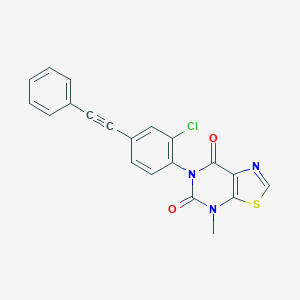 SCHEMBL16599112 SCHEMBL16599112 | C20H12ClN3O2S | 393.845 | 4 / 0 | 4.7 | Yes |
| 467722 | 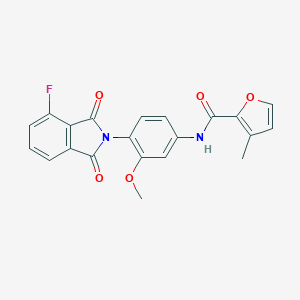 CHEMBL3634432 CHEMBL3634432 | C21H15FN2O5 | 394.358 | 6 / 1 | 3.1 | Yes |
| 39676 | 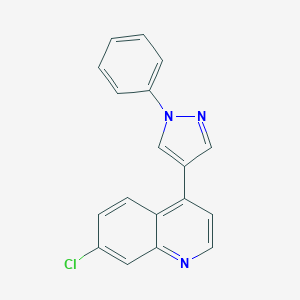 CHEMBL2023456 CHEMBL2023456 | C18H12ClN3 | 305.765 | 2 / 0 | 4.4 | Yes |
| 40143 | 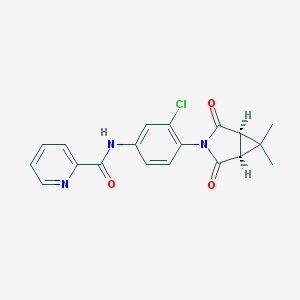 CHEMBL1711049 CHEMBL1711049 | C19H16ClN3O3 | 369.805 | 4 / 1 | 2.5 | Yes |
| 522755 | 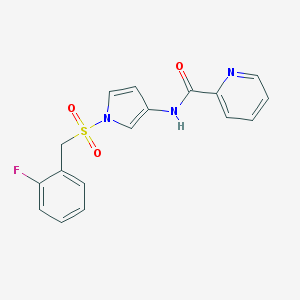 CHEMBL3809364 CHEMBL3809364 | C17H14FN3O3S | 359.375 | 5 / 1 | 2.0 | Yes |
| 467978 | 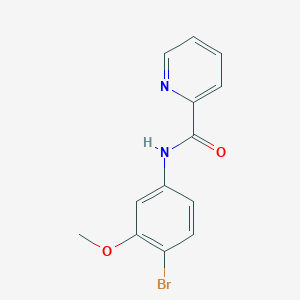 CHEMBL3609731 CHEMBL3609731 | C13H11BrN2O2 | 307.147 | 3 / 1 | 2.7 | Yes |
| 522773 | 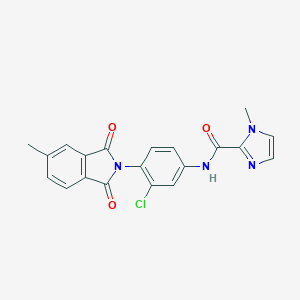 CHEMBL3759555 CHEMBL3759555 | C20H15ClN4O3 | 394.815 | 4 / 1 | 2.7 | Yes |
| 468116 | 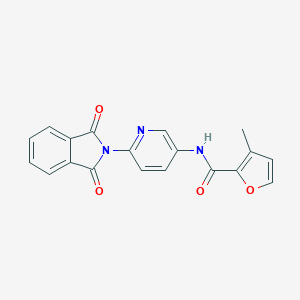 CHEMBL3634433 CHEMBL3634433 | C19H13N3O4 | 347.33 | 5 / 1 | 2.3 | Yes |
| 555639 | 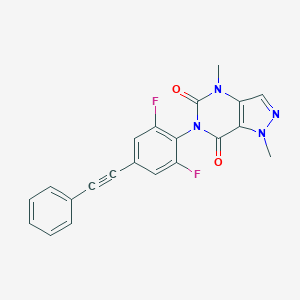 SCHEMBL16599270 SCHEMBL16599270 | C21H14F2N4O2 | 392.366 | 5 / 0 | 3.2 | Yes |
| 44932 | 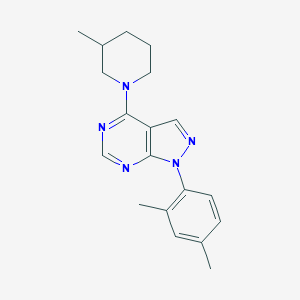 VU0080241 VU0080241 | C19H23N5 | 321.428 | 4 / 0 | 4.3 | Yes |
| 45462 | 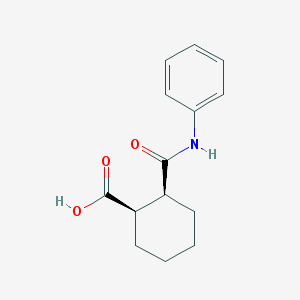 CHEMBL584209 CHEMBL584209 | C14H17NO3 | 247.294 | 3 / 2 | 2.2 | Yes |
| 459631 | 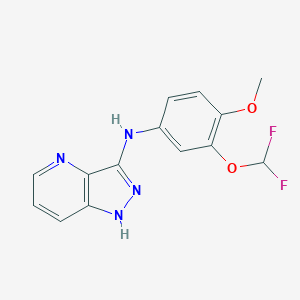 SCHEMBL6921998 SCHEMBL6921998 | C14H12F2N4O2 | 306.273 | 7 / 2 | 3.3 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417