

You can:
| Name | Cysteinyl leukotriene receptor 1 |
|---|---|
| Species | Cavia porcellus (Guinea pig) |
| Gene | CYSLTR1 |
| Synonym | CysLTR1 Cysteinyl leukotriene D4 receptor LTD4 receptor |
| Disease | N/A for non-human GPCRs |
| Length | 340 |
| Amino acid sequence | MDETGNPTIPPASNNTCYDSIDDFRNQVYSTLYSMISVVGFFGNGFVLYVLVKTYHEKSAFQVYMINLAVADLLCVCTLPLRVAYYVHKGIWLFGDFLCRLSTYALYVNLYCSIFFMTAMSFFRCVAIVFPVQNISLVTQKKARLVCIAIWMFVILTSSPFLMANTYKDEKNNTKCFEPPQDNQAKNYVLILHYVSLFIGFIIPFITIIVCYTMIIFTLLKSSMKKNLSSRKRAIGMIIVVTAAFLVSFMPYHIQRTIHLHFLHNKTKPCDSILRMQKSVVITLSLAASNCCFDPLLYFFSGGNFRRRLSTIRKYSLSSMTYIPKKKTSLPQKGKDICKE |
| UniProt | Q2NNR5 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL5645 |
| IUPHAR | N/A |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 649 | 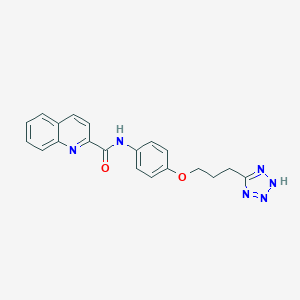 CHEMBL18388 CHEMBL18388 | C20H18N6O2 | 374.404 | 6 / 2 | 3.2 | Yes |
| 1050 | 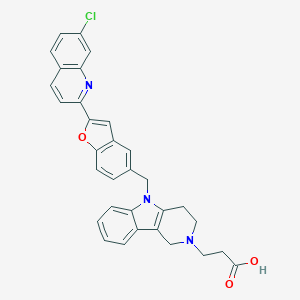 CHEMBL559928 CHEMBL559928 | C32H26ClN3O3 | 536.028 | 5 / 1 | 3.7 | No |
| 1196 | 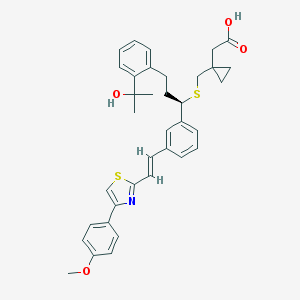 CHEMBL40836 CHEMBL40836 | C36H39NO4S2 | 613.831 | 7 / 2 | 7.5 | No |
| 3332 | 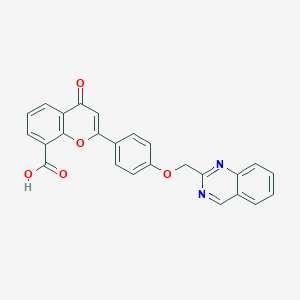 CHEMBL417135 CHEMBL417135 | C25H16N2O5 | 424.412 | 7 / 1 | 3.8 | Yes |
| 463512 | 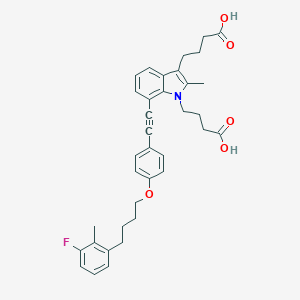 CHEMBL3597633 CHEMBL3597633 | C36H38FNO5 | 583.7 | 6 / 2 | 7.4 | No |
| 5814 | 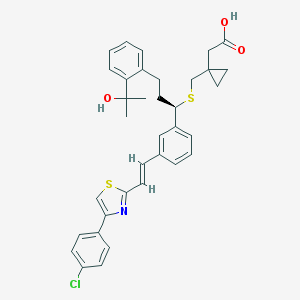 CHEMBL43821 CHEMBL43821 | C35H36ClNO3S2 | 618.247 | 6 / 2 | 8.1 | No |
| 5900 | 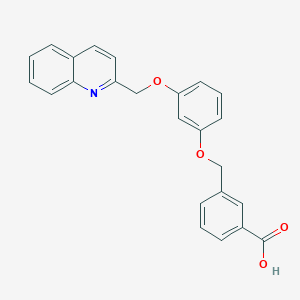 CHEMBL18979 CHEMBL18979 | C24H19NO4 | 385.419 | 5 / 1 | 4.7 | Yes |
| 7308 | 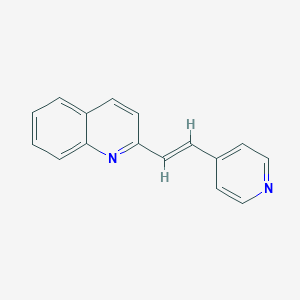 CHEMBL338876 CHEMBL338876 | C16H12N2 | 232.286 | 2 / 0 | 3.4 | Yes |
| 7373 | 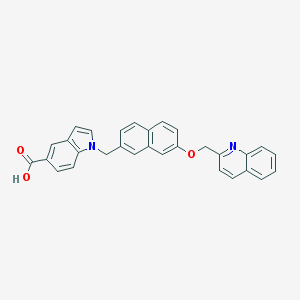 CHEMBL129079 CHEMBL129079 | C30H22N2O3 | 458.517 | 4 / 1 | 6.1 | No |
| 7514 | 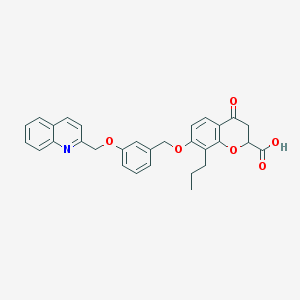 CHEMBL49325 CHEMBL49325 | C30H27NO6 | 497.547 | 7 / 1 | 5.6 | No |
| 8698 | 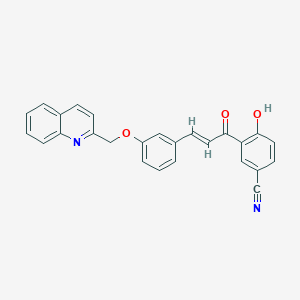 CHEMBL11820 CHEMBL11820 | C26H18N2O3 | 406.441 | 5 / 1 | 5.4 | No |
| 11214 | 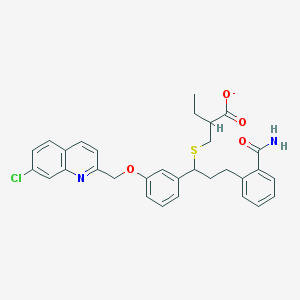 CHEMBL15120 CHEMBL15120 | C31H30ClN2O4S- | 562.101 | 6 / 1 | 7.1 | No |
| 11572 | 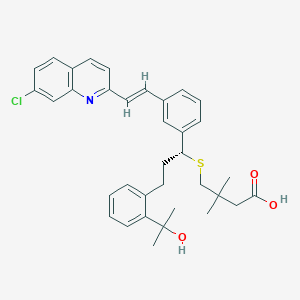 Montelukast Gem-dimethylmethylene Analogue Montelukast Gem-dimethylmethylene Analogue | C35H38ClNO3S | 588.203 | 5 / 2 | 8.3 | No |
| 12929 | 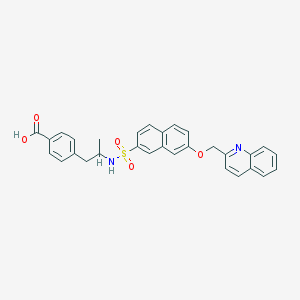 CHEMBL335000 CHEMBL335000 | C30H26N2O5S | 526.607 | 7 / 2 | 5.8 | No |
| 13189 | 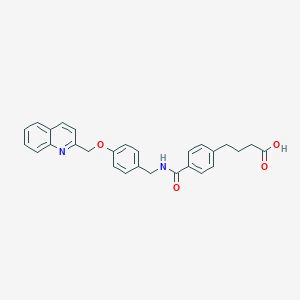 CHEMBL128151 CHEMBL128151 | C28H26N2O4 | 454.526 | 5 / 2 | 4.6 | Yes |
| 13501 | 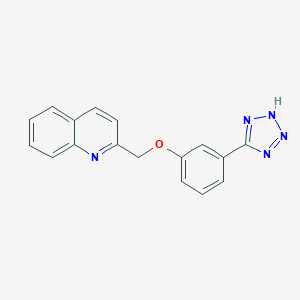 CHEMBL18864 CHEMBL18864 | C17H13N5O | 303.325 | 5 / 1 | 2.9 | Yes |
| 14203 | 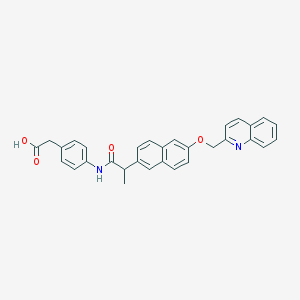 CHEMBL129866 CHEMBL129866 | C31H26N2O4 | 490.559 | 5 / 2 | 5.7 | No |
| 14223 | 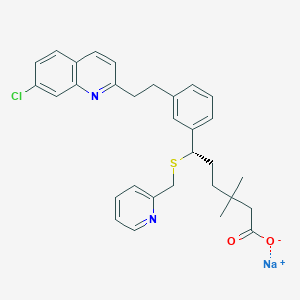 CHEMBL274038 CHEMBL274038 | C31H32ClN2NaO2S | 555.109 | 5 / 0 | N/A | No |
| 15354 | 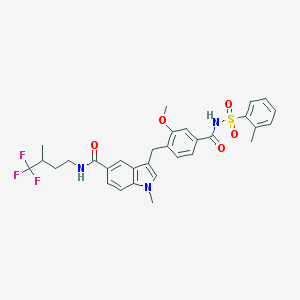 CHEMBL26284 CHEMBL26284 | C31H32F3N3O5S | 615.668 | 8 / 2 | 6.2 | No |
| 16042 | 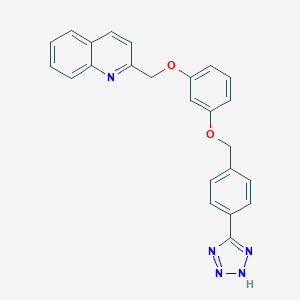 CHEMBL418002 CHEMBL418002 | C24H19N5O2 | 409.449 | 6 / 1 | 4.4 | Yes |
| 464919 | 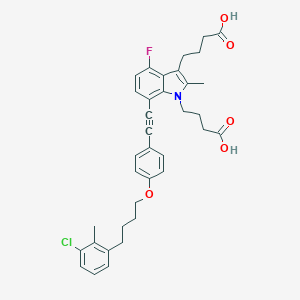 CHEMBL3597628 CHEMBL3597628 | C36H37ClFNO5 | 618.142 | 6 / 2 | 8.1 | No |
| 16790 | 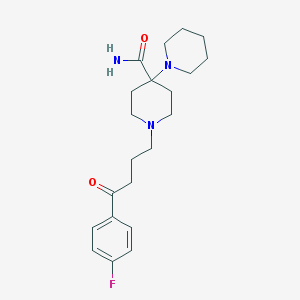 Pipamperone Pipamperone | C21H30FN3O2 | 375.488 | 5 / 1 | 2.0 | Yes |
| 17091 | 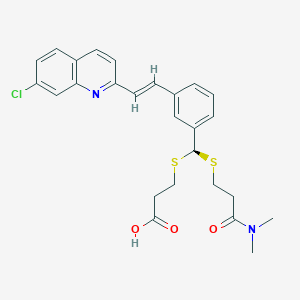 CHEMBL97257 CHEMBL97257 | C26H27ClN2O3S2 | 515.083 | 6 / 1 | 5.3 | No |
| 17094 | 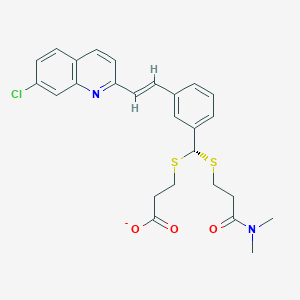 CHEMBL89340 CHEMBL89340 | C26H26ClN2O3S2- | 514.075 | 6 / 0 | 5.9 | No |
| 17095 | 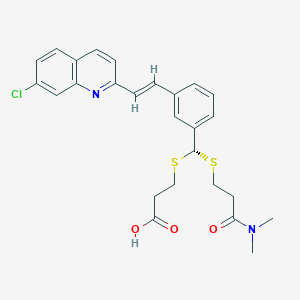 VERLUKAST VERLUKAST | C26H27ClN2O3S2 | 515.083 | 6 / 1 | 5.3 | No |
| 17099 | 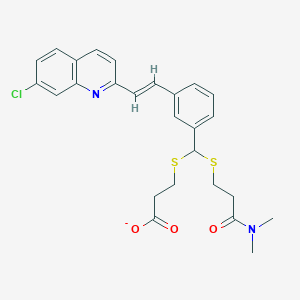 CHEMBL89768 CHEMBL89768 | C26H26ClN2O3S2- | 514.075 | 6 / 0 | 5.9 | No |
| 17100 | 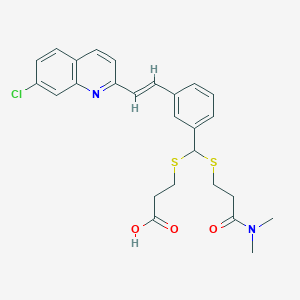 MK 571 MK 571 | C26H27ClN2O3S2 | 515.083 | 6 / 1 | 5.3 | No |
| 17271 | 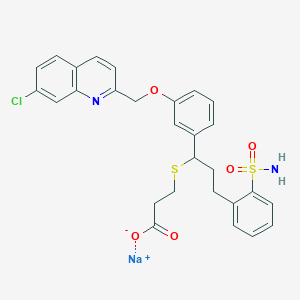 CHEMBL15292 CHEMBL15292 | C28H26ClN2NaO5S2 | 593.085 | 8 / 1 | N/A | No |
| 19362 | 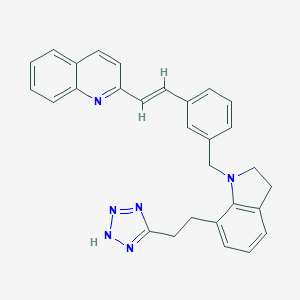 CHEMBL84150 CHEMBL84150 | C29H26N6 | 458.569 | 5 / 1 | 5.9 | No |
| 19556 | 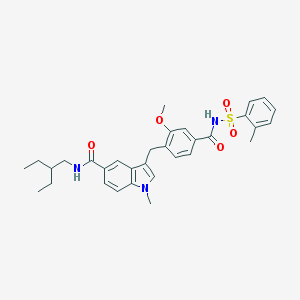 CHEMBL281066 CHEMBL281066 | C32H37N3O5S | 575.724 | 5 / 2 | 6.0 | No |
| 22272 | 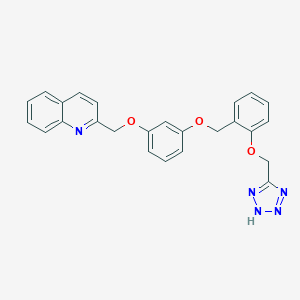 CHEMBL19072 CHEMBL19072 | C25H21N5O3 | 439.475 | 7 / 1 | 4.2 | Yes |
| 22447 | 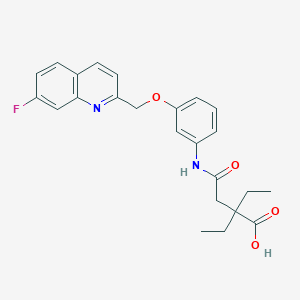 CGP-57698 CGP-57698 | C24H25FN2O4 | 424.472 | 6 / 2 | 4.0 | Yes |
| 23481 |  CHEMBL429975 CHEMBL429975 | C38H36ClN2O6S2- | 716.284 | 8 / 1 | 9.0 | No |
| 25761 |  CHEMBL337473 CHEMBL337473 | C34H39N3O5S | 601.762 | 5 / 1 | 6.5 | No |
| 26347 |  CHEMBL38052 CHEMBL38052 | C31H34O7S | 550.666 | 8 / 3 | 6.0 | No |
| 27620 |  CHEMBL131148 CHEMBL131148 | C33H39N3O5S | 589.751 | 5 / 2 | 6.5 | No |
| 28095 | 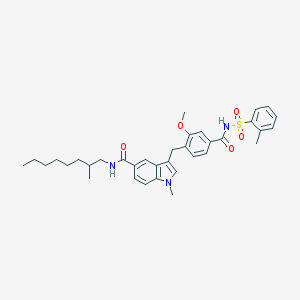 CHEMBL131173 CHEMBL131173 | C35H43N3O5S | 617.805 | 5 / 2 | 7.8 | No |
| 28523 | 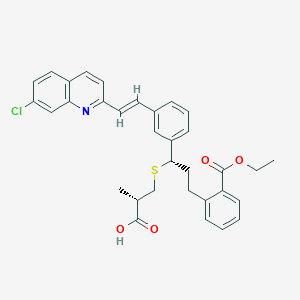 CHEMBL344756 CHEMBL344756 | C33H32ClNO4S | 574.132 | 6 / 1 | 8.2 | No |
| 31118 | 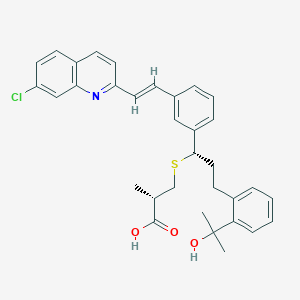 CHEMBL143874 CHEMBL143874 | C33H34ClNO3S | 560.149 | 5 / 2 | 7.7 | No |
| 31437 | 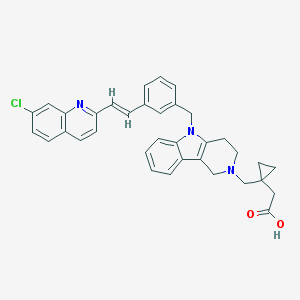 CHEMBL554154 CHEMBL554154 | C35H32ClN3O2 | 562.11 | 4 / 1 | 4.5 | No |
| 32303 | 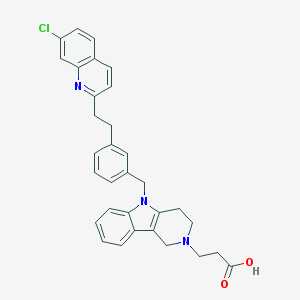 CHEMBL562575 CHEMBL562575 | C32H30ClN3O2 | 524.061 | 4 / 1 | 3.9 | No |
| 32480 | 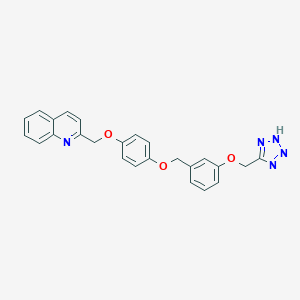 CHEMBL18956 CHEMBL18956 | C25H21N5O3 | 439.475 | 7 / 1 | 4.2 | Yes |
| 33461 | 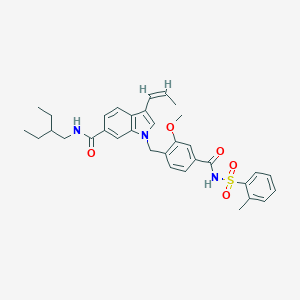 CHEMBL131514 CHEMBL131514 | C34H39N3O5S | 601.762 | 5 / 2 | 6.7 | No |
| 34492 | 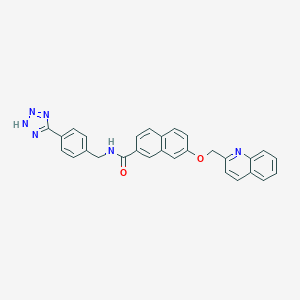 CHEMBL128477 CHEMBL128477 | C29H22N6O2 | 486.535 | 6 / 2 | 4.9 | Yes |
| 34668 |  CHEMBL32944 CHEMBL32944 | C27H17NO4 | 419.436 | 5 / 1 | 5.3 | No |
| 35163 |  CHEMBL32764 CHEMBL32764 | C27H17NO4 | 419.436 | 5 / 1 | 5.3 | No |
| 36596 |  Cinalukast Cinalukast | C23H28N2O3S | 412.548 | 5 / 2 | 4.6 | Yes |
| 36625 |  CHEMBL287559 CHEMBL287559 | C24H14BrNO5S | 508.342 | 7 / 1 | 5.4 | No |
| 37131 | 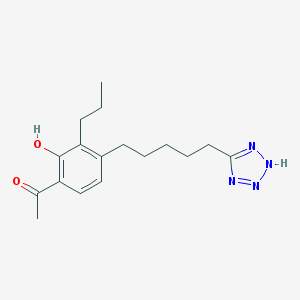 CHEMBL48483 CHEMBL48483 | C17H24N4O2 | 316.405 | 5 / 2 | 4.4 | Yes |
| 37633 | 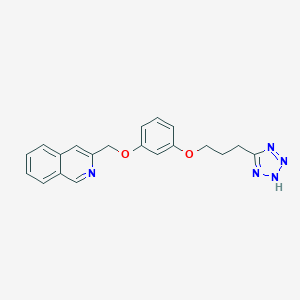 CHEMBL17973 CHEMBL17973 | C20H19N5O2 | 361.405 | 6 / 1 | 3.5 | Yes |
| 38865 | 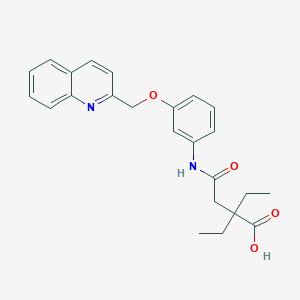 CHEMBL367424 CHEMBL367424 | C24H26N2O4 | 406.482 | 5 / 2 | 3.9 | Yes |
| 40812 | 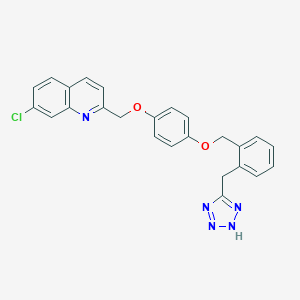 CHEMBL19158 CHEMBL19158 | C25H20ClN5O2 | 457.918 | 6 / 1 | 5.3 | No |
| 41916 | 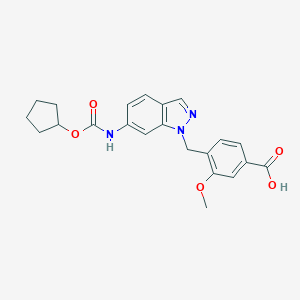 CHEMBL49298 CHEMBL49298 | C22H23N3O5 | 409.442 | 6 / 2 | 3.5 | Yes |
| 42146 | 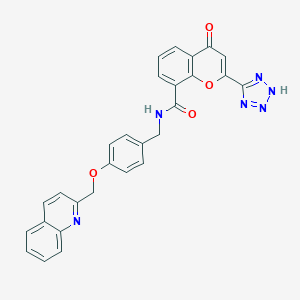 CHEMBL131287 CHEMBL131287 | C28H20N6O4 | 504.506 | 8 / 2 | 3.3 | No |
| 42394 | 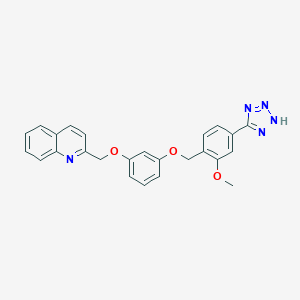 CHEMBL18589 CHEMBL18589 | C25H21N5O3 | 439.475 | 7 / 1 | 4.3 | Yes |
| 42955 | 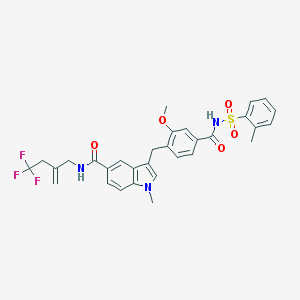 CHEMBL26332 CHEMBL26332 | C31H30F3N3O5S | 613.652 | 8 / 2 | 6.2 | No |
| 44529 | 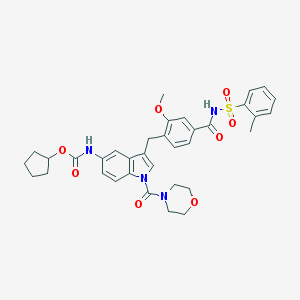 CHEMBL83166 CHEMBL83166 | C35H38N4O8S | 674.769 | 8 / 2 | 5.0 | No |
| 45083 | 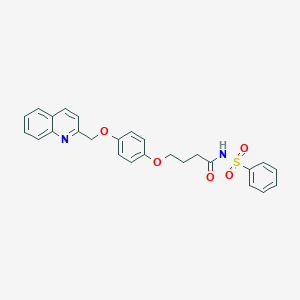 CHEMBL278920 CHEMBL278920 | C26H24N2O5S | 476.547 | 6 / 1 | 4.4 | Yes |
| 46061 | 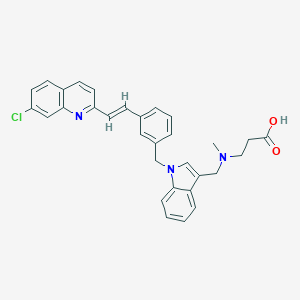 CHEMBL563165 CHEMBL563165 | C31H28ClN3O2 | 510.034 | 4 / 1 | 3.8 | No |
| 47602 | 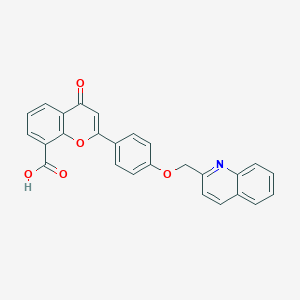 CHEMBL33294 CHEMBL33294 | C26H17NO5 | 423.424 | 6 / 1 | 4.5 | Yes |
| 51359 | 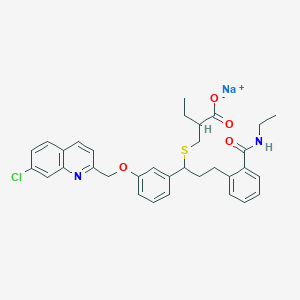 CHEMBL280310 CHEMBL280310 | C33H34ClN2NaO4S | 613.145 | 6 / 1 | N/A | No |
| 51749 | 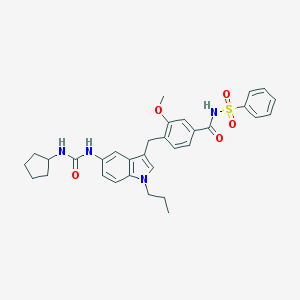 CHEMBL52492 CHEMBL52492 | C32H36N4O5S | 588.723 | 5 / 3 | 5.4 | No |
| 51767 | 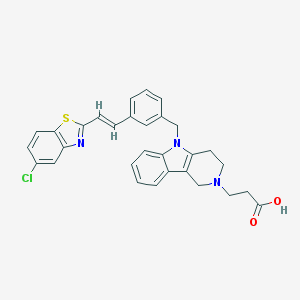 CHEMBL565228 CHEMBL565228 | C30H26ClN3O2S | 528.067 | 5 / 1 | 4.1 | No |
| 51768 | 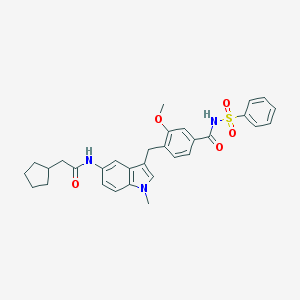 CHEMBL50370 CHEMBL50370 | C31H33N3O5S | 559.681 | 5 / 2 | 5.6 | No |
| 52204 | 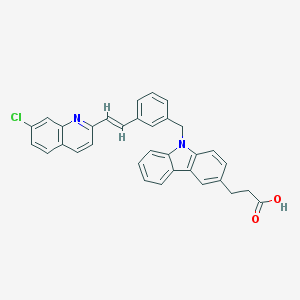 CHEMBL562930 CHEMBL562930 | C33H25ClN2O2 | 517.025 | 3 / 1 | 7.9 | No |
| 52723 | 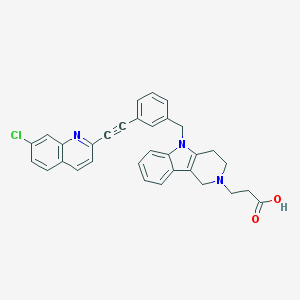 CHEMBL562383 CHEMBL562383 | C32H26ClN3O2 | 520.029 | 4 / 1 | 3.7 | No |
| 54533 | 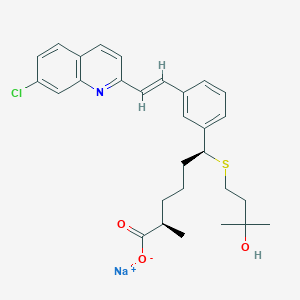 CHEMBL416344 CHEMBL416344 | C29H33ClNNaO3S | 534.087 | 5 / 1 | N/A | No |
| 54534 | 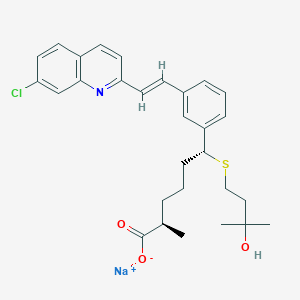 CHEMBL275823 CHEMBL275823 | C29H33ClNNaO3S | 534.087 | 5 / 1 | N/A | No |
| 55924 | 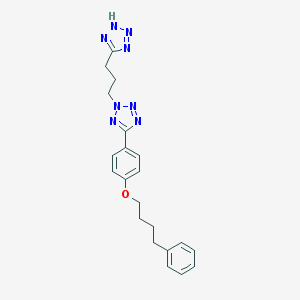 CHEMBL19085 CHEMBL19085 | C21H24N8O | 404.478 | 7 / 1 | 4.3 | Yes |
| 56380 | 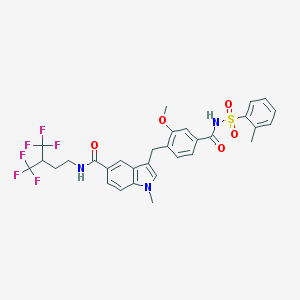 CHEMBL24505 CHEMBL24505 | C31H29F6N3O5S | 669.639 | 11 / 2 | 6.8 | No |
| 56900 | 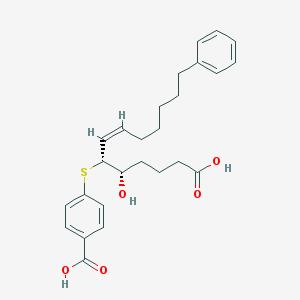 CHEMBL39586 CHEMBL39586 | C26H32O5S | 456.597 | 6 / 3 | 5.9 | No |
| 58118 | 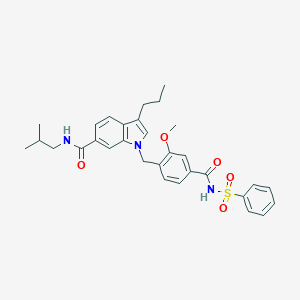 CHEMBL422616 CHEMBL422616 | C31H35N3O5S | 561.697 | 5 / 2 | 5.9 | No |
| 58490 | 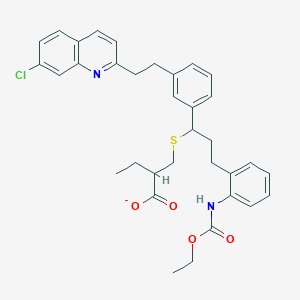 CHEMBL279630 CHEMBL279630 | C34H36ClN2O4S- | 604.182 | 6 / 1 | 8.9 | No |
| 59431 | 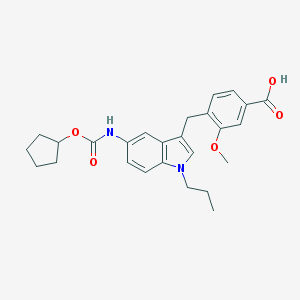 CHEMBL422404 CHEMBL422404 | C26H30N2O5 | 450.535 | 5 / 2 | 5.1 | No |
| 59828 | 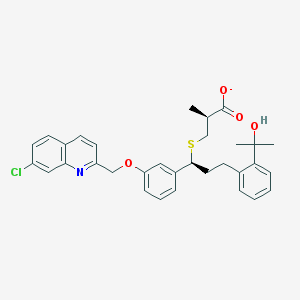 CHEMBL274913 CHEMBL274913 | C32H33ClNO4S- | 563.129 | 6 / 1 | 7.5 | No |
| 59829 | 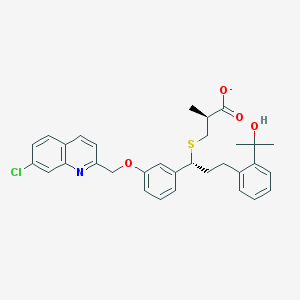 CHEMBL15306 CHEMBL15306 | C32H33ClNO4S- | 563.129 | 6 / 1 | 7.5 | No |
| 59830 | 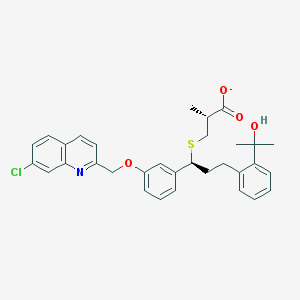 CHEMBL277533 CHEMBL277533 | C32H33ClNO4S- | 563.129 | 6 / 1 | 7.5 | No |
| 59831 | 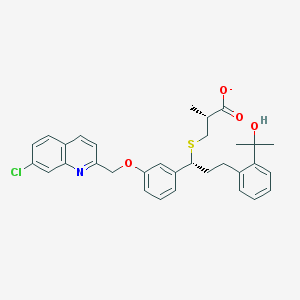 CHEMBL275357 CHEMBL275357 | C32H33ClNO4S- | 563.129 | 6 / 1 | 7.5 | No |
| 444063 | 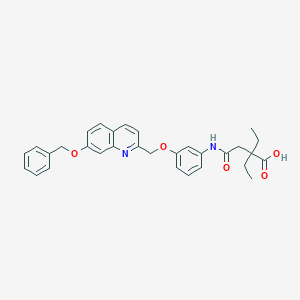 CHEMBL368841 CHEMBL368841 | C31H32N2O5 | 512.606 | 6 / 2 | 5.4 | No |
| 60426 | 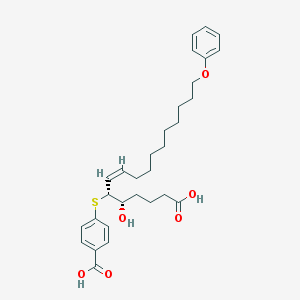 CHEMBL39794 CHEMBL39794 | C30H40O6S | 528.704 | 7 / 3 | 7.4 | No |
| 60718 | 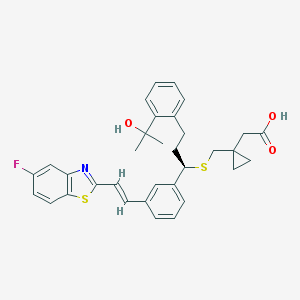 CHEMBL43509 CHEMBL43509 | C33H34FNO3S2 | 575.757 | 7 / 2 | 7.4 | No |
| 62807 | 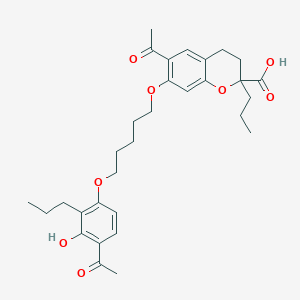 CHEMBL54744 CHEMBL54744 | C31H40O8 | 540.653 | 8 / 2 | 6.6 | No |
| 63540 | 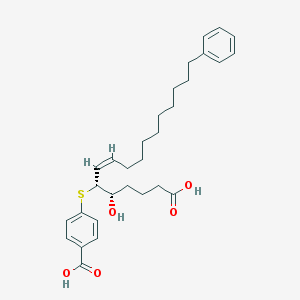 CHEMBL40009 CHEMBL40009 | C30H40O5S | 512.705 | 6 / 3 | 8.1 | No |
| 64625 | 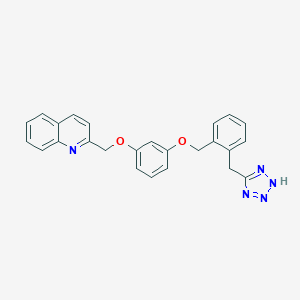 CHEMBL18204 CHEMBL18204 | C25H21N5O2 | 423.476 | 6 / 1 | 4.7 | Yes |
| 64949 | 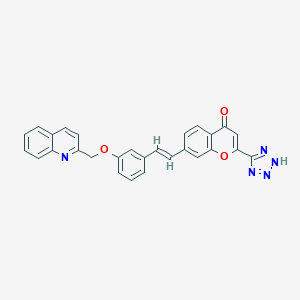 CHEMBL36113 CHEMBL36113 | C28H19N5O3 | 473.492 | 7 / 1 | 4.8 | Yes |
| 65511 | 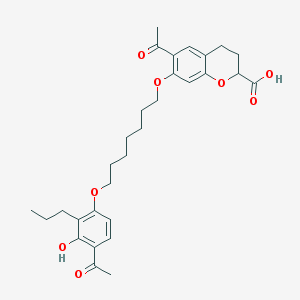 CHEMBL54542 CHEMBL54542 | C30H38O8 | 526.626 | 8 / 2 | 6.4 | No |
| 66563 | 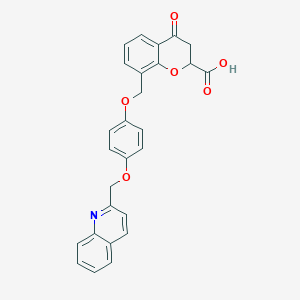 CHEMBL45965 CHEMBL45965 | C27H21NO6 | 455.466 | 7 / 1 | 4.2 | Yes |
| 67190 | 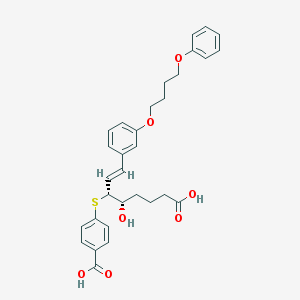 CHEMBL289540 CHEMBL289540 | C31H34O7S | 550.666 | 8 / 3 | 6.1 | No |
| 67191 | 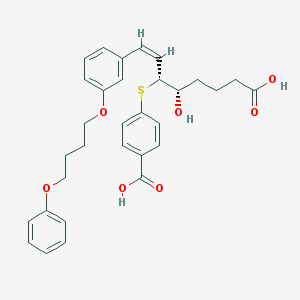 CHEMBL40153 CHEMBL40153 | C31H34O7S | 550.666 | 8 / 3 | 6.1 | No |
| 67323 | 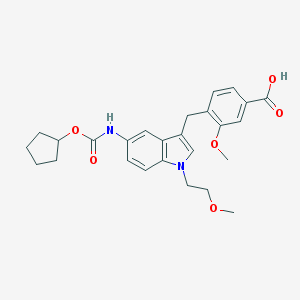 CHEMBL50422 CHEMBL50422 | C26H30N2O6 | 466.534 | 6 / 2 | 4.1 | Yes |
| 67405 | 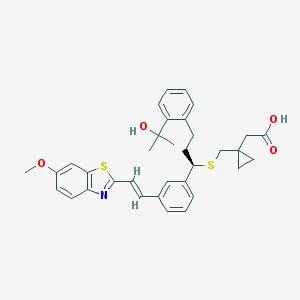 CHEMBL42410 CHEMBL42410 | C34H37NO4S2 | 587.793 | 7 / 2 | 7.3 | No |
| 69357 | 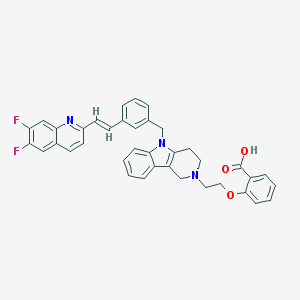 CHEMBL555048 CHEMBL555048 | C38H31F2N3O3 | 615.681 | 7 / 1 | 5.2 | No |
| 71649 | 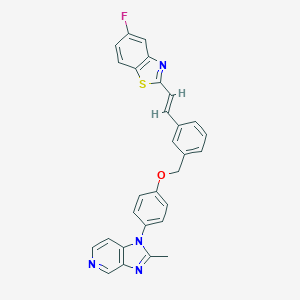 CHEMBL31714 CHEMBL31714 | C29H21FN4OS | 492.572 | 6 / 0 | 6.8 | No |
| 72465 | 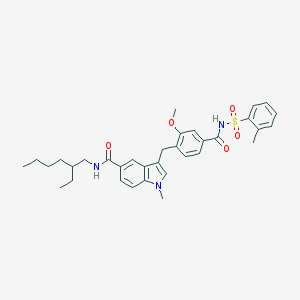 CHEMBL413742 CHEMBL413742 | C34H41N3O5S | 603.778 | 5 / 2 | 7.1 | No |
| 73662 | 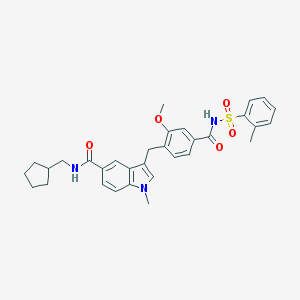 CHEMBL24256 CHEMBL24256 | C32H35N3O5S | 573.708 | 5 / 2 | 5.8 | No |
| 74147 | 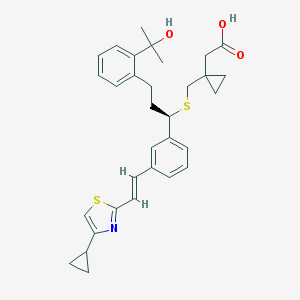 CHEMBL295281 CHEMBL295281 | C32H37NO3S2 | 547.772 | 6 / 2 | 6.4 | No |
| 75396 | 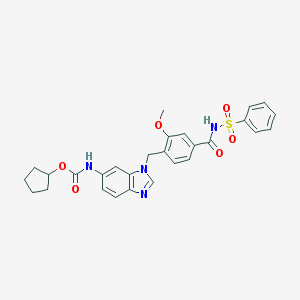 107786-78-7 107786-78-7 | C28H28N4O6S | 548.614 | 7 / 2 | 4.3 | No |
| 76099 | 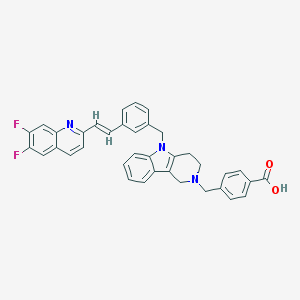 CHEMBL555910 CHEMBL555910 | C37H29F2N3O2 | 585.655 | 6 / 1 | 5.0 | No |
| 76197 | 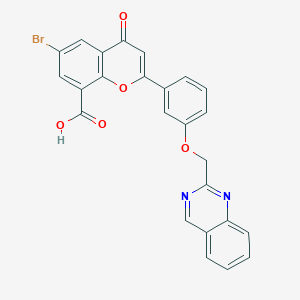 CHEMBL34740 CHEMBL34740 | C25H15BrN2O5 | 503.308 | 7 / 1 | 4.5 | No |
| 76286 | 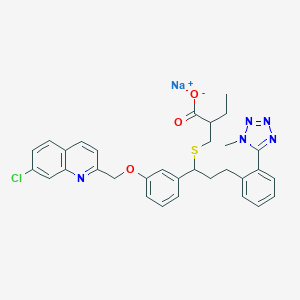 CHEMBL15232 CHEMBL15232 | C32H31ClN5NaO3S | 624.132 | 8 / 0 | N/A | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417