

You can:
| Name | Lysophosphatidic acid receptor 1 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | LPAR1 |
| Synonym | lysophosphatidic acid receptor Edg-2 Lpar1 LPA1 receptor Lysophosphatidic acid receptor Edg-2 {ECO:0000303|PubMed:9070858} LPA-1 [ Show all ] |
| Disease | Idiopathic pulmonary fibrosis |
| Length | 364 |
| Amino acid sequence | MAAISTSIPVISQPQFTAMNEPQCFYNESIAFFYNRSGKHLATEWNTVSKLVMGLGITVCIFIMLANLLVMVAIYVNRRFHFPIYYLMANLAAADFFAGLAYFYLMFNTGPNTRRLTVSTWLLRQGLIDTSLTASVANLLAIAIERHITVFRMQLHTRMSNRRVVVVIVVIWTMAIVMGAIPSVGWNCICDIENCSNMAPLYSDSYLVFWAIFNLVTFVVMVVLYAHIFGYVRQRTMRMSRHSSGPRRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCCPQCDVLAYEKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQILCCQRSENPTGPTEGSDRSASSLNHTILAGVHSNDHSVV |
| UniProt | Q92633 |
| Protein Data Bank | 4z36, 4z35, 4z34 |
| GPCR-HGmod model | Q92633 |
| 3D structure model | This structure is from PDB ID 4z36. |
| BioLiP | BL0315557, BL0315553, BL0315554, BL0315555, BL0315556, BL0315558 |
| Therapeutic Target Database | T92640 |
| ChEMBL | CHEMBL3819 |
| IUPHAR | 272 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 535926 | 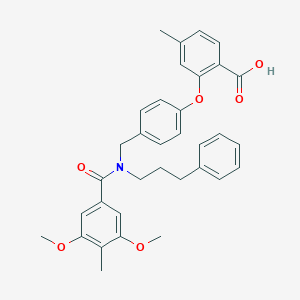 CHEMBL3917128 CHEMBL3917128 | C34H35NO6 | 553.655 | 6 / 1 | 6.9 | No |
| 535972 | 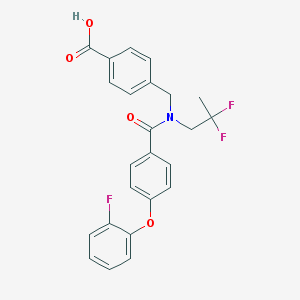 CHEMBL3943469 CHEMBL3943469 | C24H20F3NO4 | 443.422 | 7 / 1 | 5.1 | No |
| 536051 | 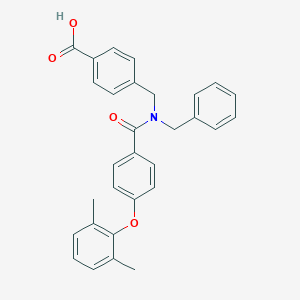 CHEMBL3920547 CHEMBL3920547 | C30H27NO4 | 465.549 | 4 / 1 | 6.2 | No |
| 15756 | 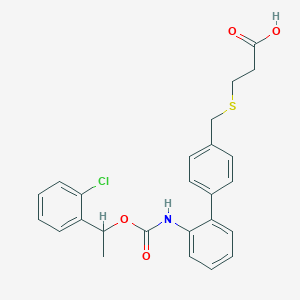 CHEMBL245895 CHEMBL245895 | C25H24ClNO4S | 469.98 | 5 / 2 | 5.6 | No |
| 536421 | 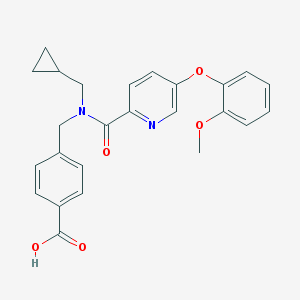 CHEMBL3985179 CHEMBL3985179 | C25H24N2O5 | 432.476 | 6 / 1 | 3.9 | Yes |
| 17531 | 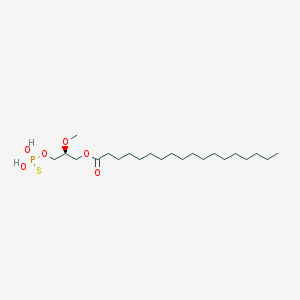 CHEMBL357053 CHEMBL357053 | C22H45O6PS | 468.63 | 7 / 2 | 8.4 | No |
| 17538 | 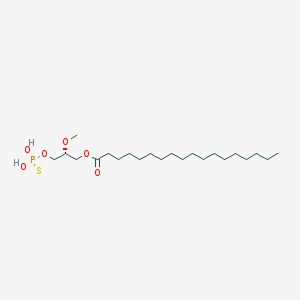 CHEMBL153043 CHEMBL153043 | C22H45O6PS | 468.63 | 7 / 2 | 8.4 | No |
| 17545 | 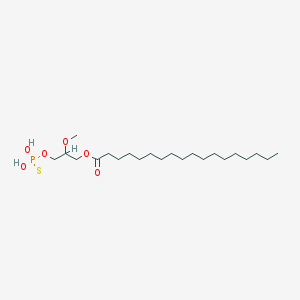 CHEMBL2017139 CHEMBL2017139 | C22H45O6PS | 468.63 | 7 / 2 | 8.4 | No |
| 19348 | 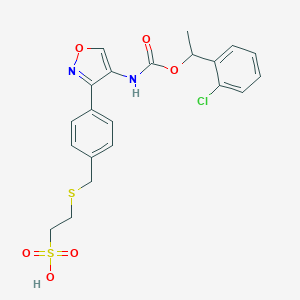 CHEMBL245295 CHEMBL245295 | C21H21ClN2O6S2 | 496.977 | 8 / 2 | 3.5 | Yes |
| 20484 | 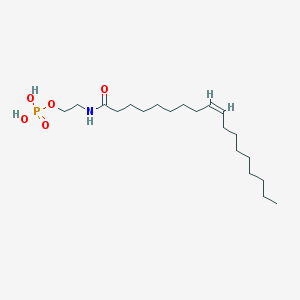 NAEPA NAEPA | C20H40NO5P | 405.516 | 5 / 3 | 5.2 | No |
| 536533 | 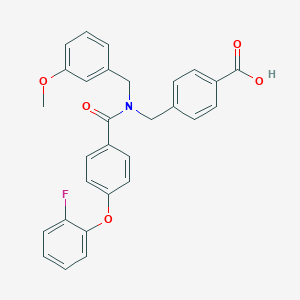 CHEMBL3900559 CHEMBL3900559 | C29H24FNO5 | 485.511 | 6 / 1 | 5.5 | No |
| 442568 | 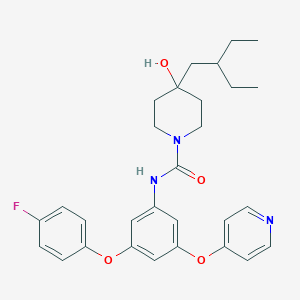 CHEMBL3401388 CHEMBL3401388 | C29H34FN3O4 | 507.606 | 6 / 2 | 5.7 | No |
| 536611 | 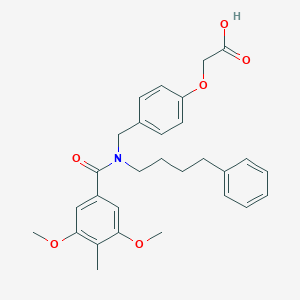 CHEMBL3953068 CHEMBL3953068 | C29H33NO6 | 491.584 | 6 / 1 | 5.4 | No |
| 536614 | 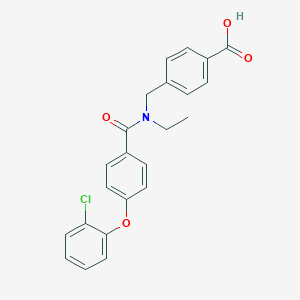 CHEMBL3942927 CHEMBL3942927 | C23H20ClNO4 | 409.866 | 4 / 1 | 4.9 | Yes |
| 536641 | 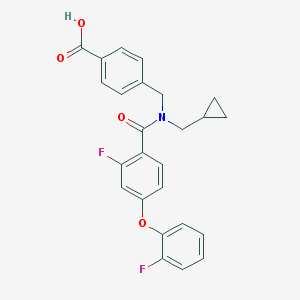 CHEMBL3900867 CHEMBL3900867 | C25H21F2NO4 | 437.443 | 6 / 1 | 4.9 | Yes |
| 25977 | 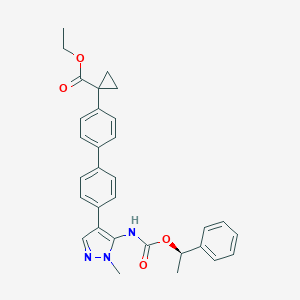 CHEMBL2182062 CHEMBL2182062 | C31H31N3O4 | 509.606 | 5 / 1 | 5.8 | No |
| 26406 | 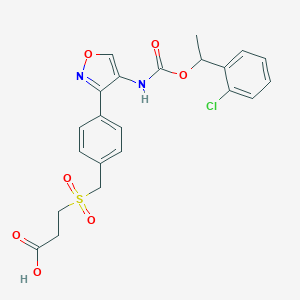 CHEMBL394038 CHEMBL394038 | C22H21ClN2O7S | 492.927 | 8 / 2 | 2.8 | Yes |
| 29257 | 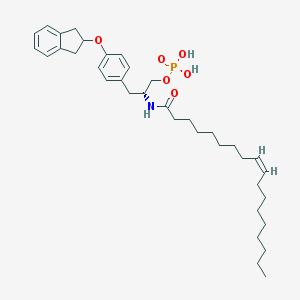 CHEMBL313413 CHEMBL313413 | C36H54NO6P | 627.803 | 6 / 3 | 9.3 | No |
| 29834 | 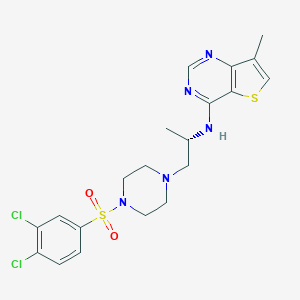 LPA2 antagonist 1 LPA2 antagonist 1 | C20H23Cl2N5O2S2 | 500.457 | 8 / 1 | 4.3 | No |
| 30464 | 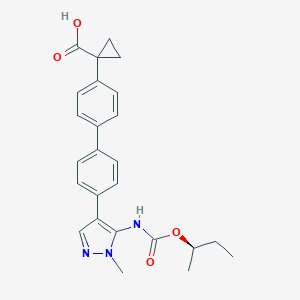 CHEMBL2182024 CHEMBL2182024 | C25H27N3O4 | 433.508 | 5 / 2 | 4.5 | Yes |
| 536814 | 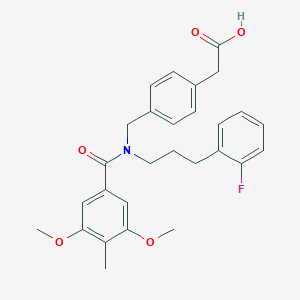 CHEMBL3944344 CHEMBL3944344 | C28H30FNO5 | 479.548 | 6 / 1 | 5.1 | No |
| 467011 | 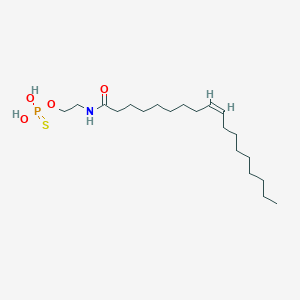 CHEMBL3621959 CHEMBL3621959 | C20H40NO4PS | 421.577 | 5 / 3 | 7.0 | No |
| 33970 | 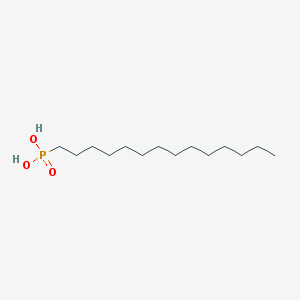 Tetradecylphosphonic acid Tetradecylphosphonic acid | C14H31O3P | 278.373 | 3 / 2 | 5.1 | No |
| 34951 | 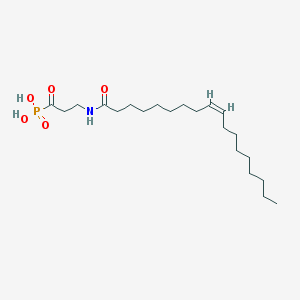 CHEMBL117529 CHEMBL117529 | C21H40NO5P | 417.527 | 5 / 3 | 4.9 | Yes |
| 36916 | 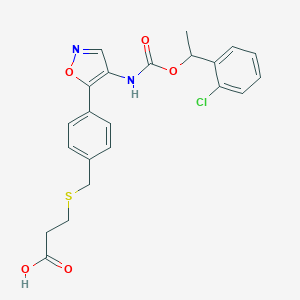 CHEMBL246928 CHEMBL246928 | C22H21ClN2O5S | 460.929 | 7 / 2 | 4.1 | Yes |
| 548354 | 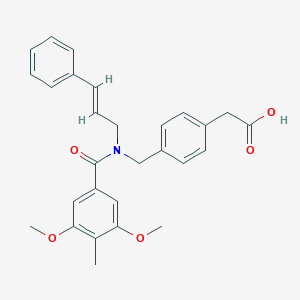 CHEMBL3973793 CHEMBL3973793 | C28H29NO5 | 459.542 | 5 / 1 | 4.8 | Yes |
| 43193 | 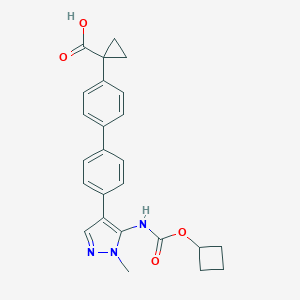 CHEMBL2182026 CHEMBL2182026 | C25H25N3O4 | 431.492 | 5 / 2 | 4.1 | Yes |
| 443440 | 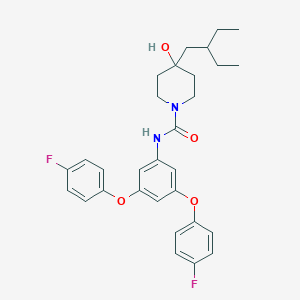 CHEMBL3401379 CHEMBL3401379 | C30H34F2N2O4 | 524.609 | 6 / 2 | 6.8 | No |
| 537152 | 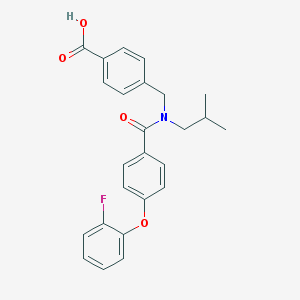 CHEMBL3948456 CHEMBL3948456 | C25H24FNO4 | 421.468 | 5 / 1 | 5.4 | No |
| 47584 | 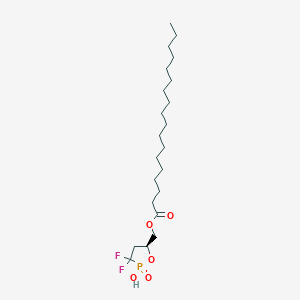 CHEMBL215722 CHEMBL215722 | C22H41F2O5P | 454.536 | 7 / 1 | 8.2 | No |
| 47627 | 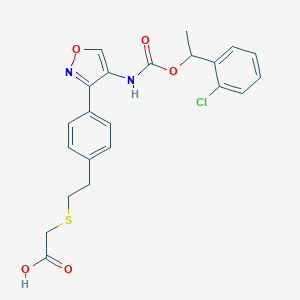 CHEMBL246942 CHEMBL246942 | C22H21ClN2O5S | 460.929 | 7 / 2 | 4.6 | Yes |
| 548453 | 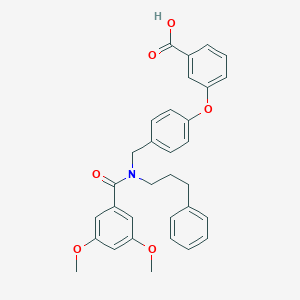 CHEMBL3956293 CHEMBL3956293 | C32H31NO6 | 525.601 | 6 / 1 | 6.2 | No |
| 49424 | 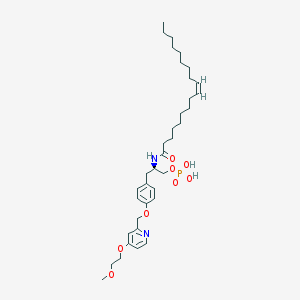 CHEMBL182446 CHEMBL182446 | C36H57N2O8P | 676.832 | 9 / 3 | 7.5 | No |
| 49614 | 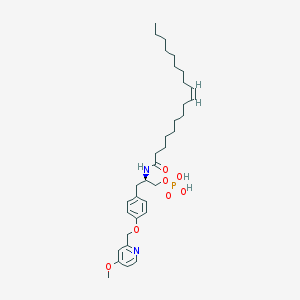 CHEMBL265967 CHEMBL265967 | C34H53N2O7P | 632.779 | 8 / 3 | 7.7 | No |
| 50273 | 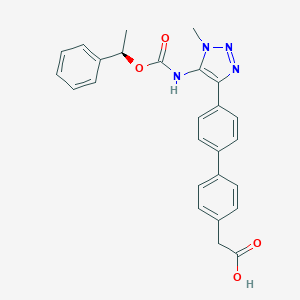 CHEMBL2182042 CHEMBL2182042 | C26H24N4O4 | 456.502 | 6 / 2 | 4.2 | Yes |
| 51249 | 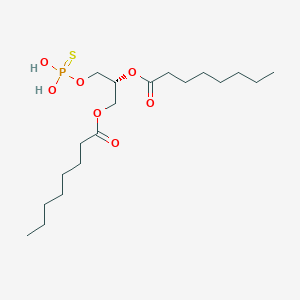 CHEMBL202361 CHEMBL202361 | C19H37O7PS | 440.532 | 8 / 2 | 6.0 | No |
| 537350 | 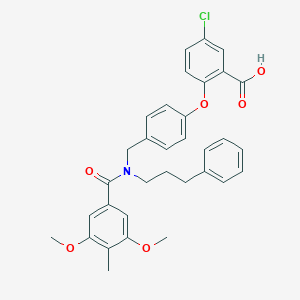 CHEMBL3975893 CHEMBL3975893 | C33H32ClNO6 | 574.07 | 6 / 1 | 7.2 | No |
| 537380 | 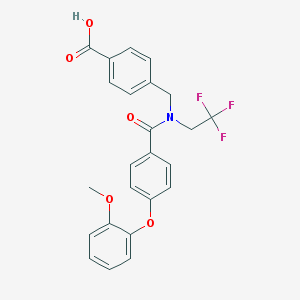 CHEMBL3901720 CHEMBL3901720 | C24H20F3NO5 | 459.421 | 8 / 1 | 5.0 | Yes |
| 537396 | 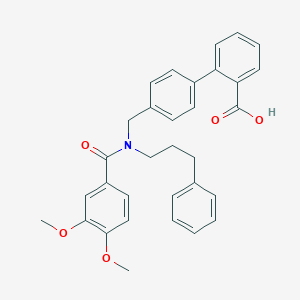 CHEMBL3979511 CHEMBL3979511 | C32H31NO5 | 509.602 | 5 / 1 | 6.3 | No |
| 57809 | 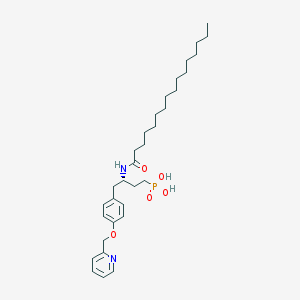 CHEMBL360064 CHEMBL360064 | C32H51N2O5P | 574.743 | 6 / 3 | 7.9 | No |
| 60441 | 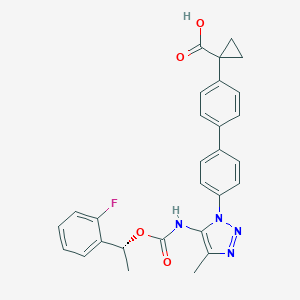 CHEMBL2182030 CHEMBL2182030 | C28H25FN4O4 | 500.53 | 7 / 2 | 5.1 | No |
| 65692 | 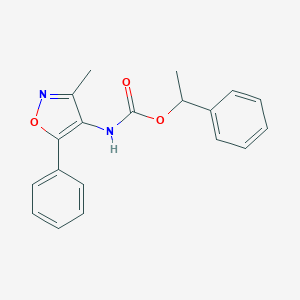 CHEMBL404575 CHEMBL404575 | C19H18N2O3 | 322.364 | 4 / 1 | 3.9 | Yes |
| 67558 | 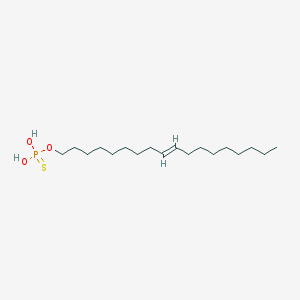 CHEMBL187459 CHEMBL187459 | C18H37O3PS | 364.525 | 4 / 2 | 8.1 | No |
| 71929 | 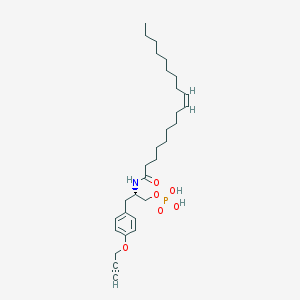 CHEMBL328474 CHEMBL328474 | C30H48NO6P | 549.689 | 6 / 3 | 7.3 | No |
| 537836 | 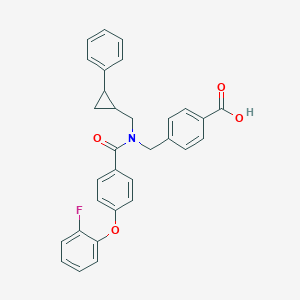 CHEMBL3980736 CHEMBL3980736 | C31H26FNO4 | 495.55 | 5 / 1 | 6.2 | No |
| 537838 | 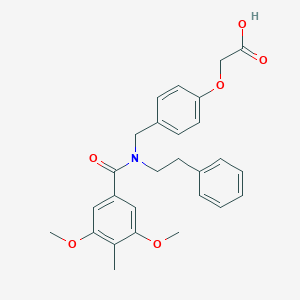 CHEMBL3981813 CHEMBL3981813 | C27H29NO6 | 463.53 | 6 / 1 | 4.7 | Yes |
| 472300 | 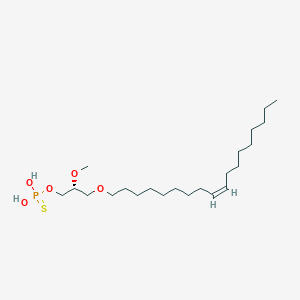 CHEMBL3621961 CHEMBL3621961 | C22H45O5PS | 452.631 | 6 / 2 | 7.8 | No |
| 76231 | 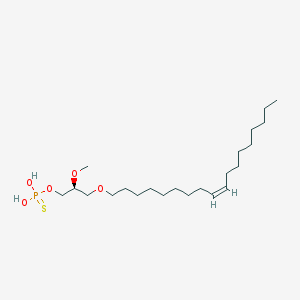 alkyl OMPT alkyl OMPT | C22H45O5PS | 452.631 | 6 / 2 | 7.8 | No |
| 77230 | 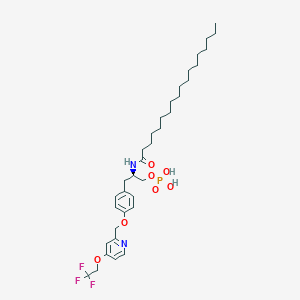 CHEMBL3218459 CHEMBL3218459 | C35H54F3N2O7P | 702.793 | 11 / 3 | 9.7 | No |
| 77520 | 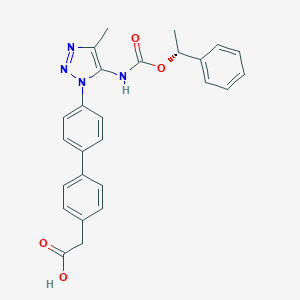 CHEMBL2182028 CHEMBL2182028 | C26H24N4O4 | 456.502 | 6 / 2 | 4.6 | Yes |
| 79844 | 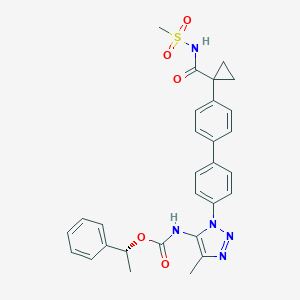 CHEMBL2182037 CHEMBL2182037 | C29H29N5O5S | 559.641 | 7 / 2 | 4.3 | No |
| 538060 | 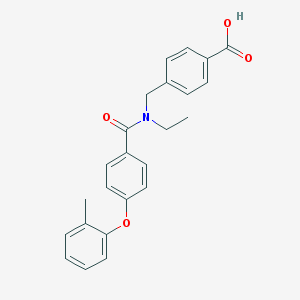 CHEMBL3908939 CHEMBL3908939 | C24H23NO4 | 389.451 | 4 / 1 | 4.7 | Yes |
| 538114 | 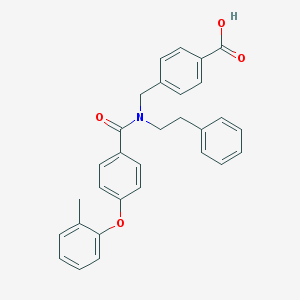 CHEMBL3971670 CHEMBL3971670 | C30H27NO4 | 465.549 | 4 / 1 | 6.3 | No |
| 86134 | 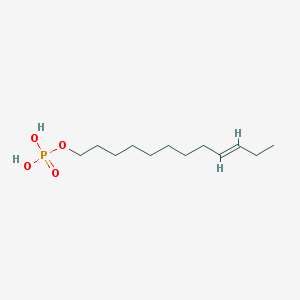 CHEMBL190349 CHEMBL190349 | C12H25O4P | 264.302 | 4 / 2 | 3.1 | Yes |
| 86328 | 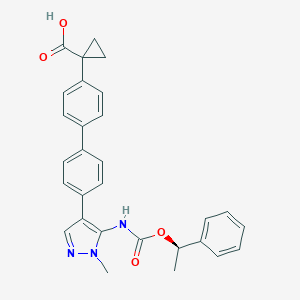 CHEMBL2182063 CHEMBL2182063 | C29H27N3O4 | 481.552 | 5 / 2 | 5.1 | No |
| 86862 | 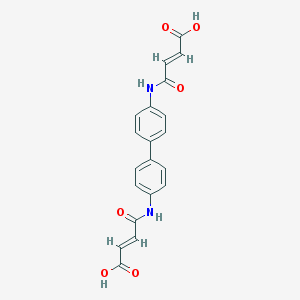 36840-10-5 36840-10-5 | C20H16N2O6 | 380.356 | 6 / 4 | 1.4 | Yes |
| 538167 | 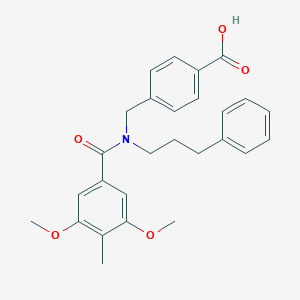 CHEMBL3905099 CHEMBL3905099 | C27H29NO5 | 447.531 | 5 / 1 | 5.0 | Yes |
| 553699 | 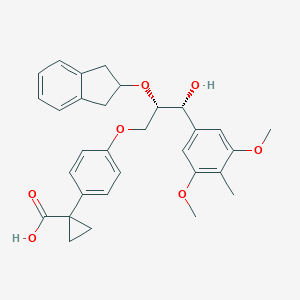 ONO-3080573 ONO-3080573 | C31H34O7 | 518.606 | 7 / 2 | 4.8 | No |
| 89073 | 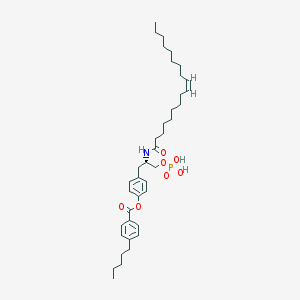 CHEMBL314553 CHEMBL314553 | C39H60NO7P | 685.883 | 7 / 3 | 11.1 | No |
| 89770 | 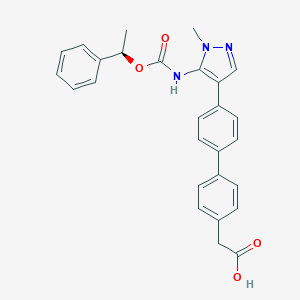 CHEMBL2182061 CHEMBL2182061 | C27H25N3O4 | 455.514 | 5 / 2 | 4.8 | Yes |
| 91234 | 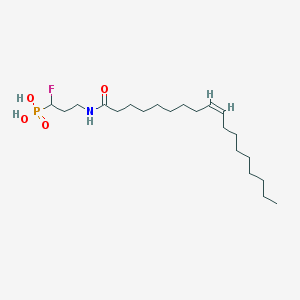 CHEMBL117663 CHEMBL117663 | C21H41FNO4P | 421.534 | 5 / 3 | 6.1 | No |
| 93762 | 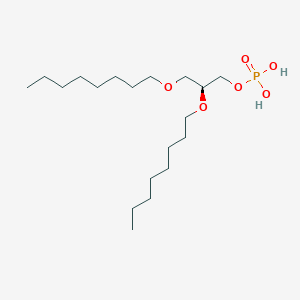 CHEMBL203986 CHEMBL203986 | C19H41O6P | 396.505 | 6 / 2 | 5.1 | No |
| 93764 | 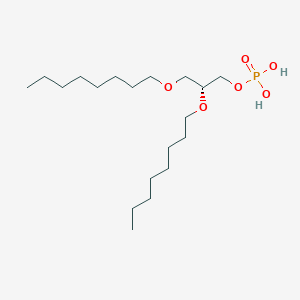 CHEMBL202243 CHEMBL202243 | C19H41O6P | 396.505 | 6 / 2 | 5.1 | No |
| 538362 | 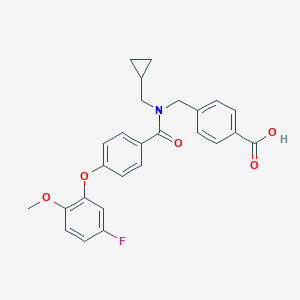 CHEMBL3921376 CHEMBL3921376 | C26H24FNO5 | 449.478 | 6 / 1 | 4.7 | Yes |
| 96492 | 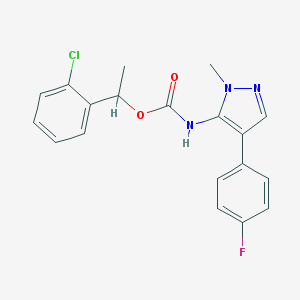 CHEMBL2182057 CHEMBL2182057 | C19H17ClFN3O2 | 373.812 | 4 / 1 | 4.4 | Yes |
| 96550 | 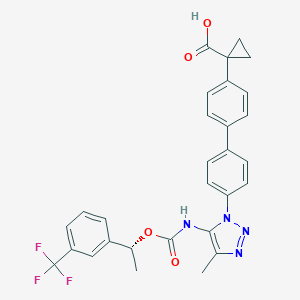 CHEMBL2182032 CHEMBL2182032 | C29H25F3N4O4 | 550.538 | 9 / 2 | 5.9 | No |
| 100156 | 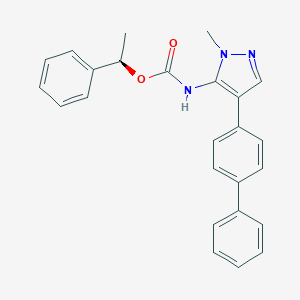 CHEMBL2182055 CHEMBL2182055 | C25H23N3O2 | 397.478 | 3 / 1 | 5.3 | No |
| 102484 | 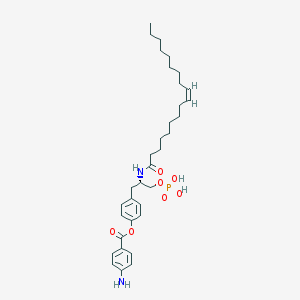 CHEMBL94245 CHEMBL94245 | C34H51N2O7P | 630.763 | 8 / 4 | 8.0 | No |
| 445758 | 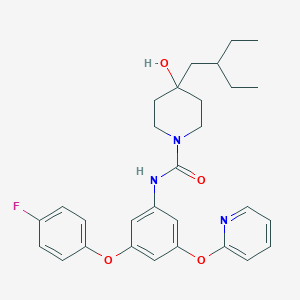 CHEMBL3401390 CHEMBL3401390 | C29H34FN3O4 | 507.606 | 6 / 2 | 6.0 | No |
| 538551 | 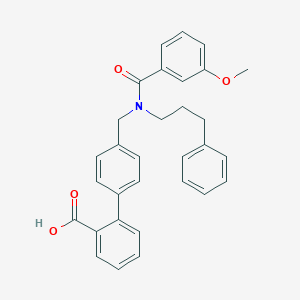 CHEMBL3915800 CHEMBL3915800 | C31H29NO4 | 479.576 | 4 / 1 | 6.3 | No |
| 104952 | 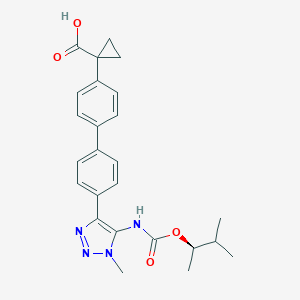 CHEMBL2182045 CHEMBL2182045 | C25H28N4O4 | 448.523 | 6 / 2 | 4.5 | Yes |
| 538579 | 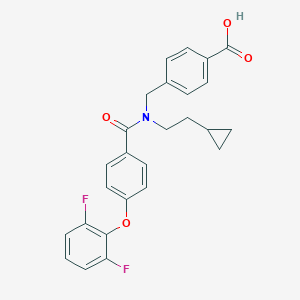 CHEMBL3893992 CHEMBL3893992 | C26H23F2NO4 | 451.47 | 6 / 1 | 5.6 | No |
| 538601 | 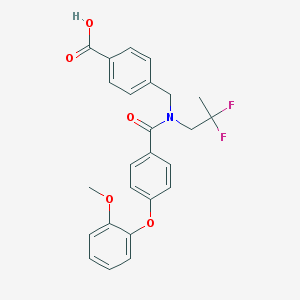 CHEMBL3892760 CHEMBL3892760 | C25H23F2NO5 | 455.458 | 7 / 1 | 4.9 | Yes |
| 538645 | 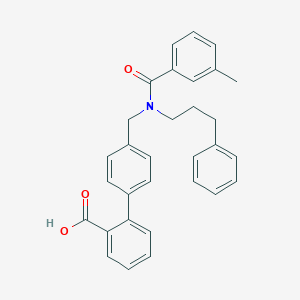 CHEMBL3902849 CHEMBL3902849 | C31H29NO3 | 463.577 | 3 / 1 | 6.7 | No |
| 538673 | 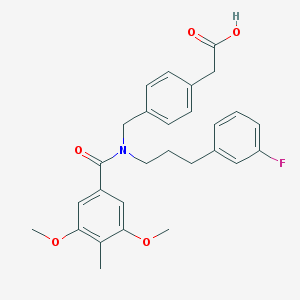 CHEMBL3901754 CHEMBL3901754 | C28H30FNO5 | 479.548 | 6 / 1 | 5.1 | No |
| 538729 | 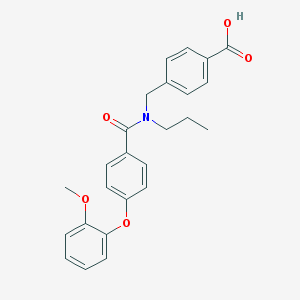 CHEMBL3961765 CHEMBL3961765 | C25H25NO5 | 419.477 | 5 / 1 | 4.8 | Yes |
| 113882 | 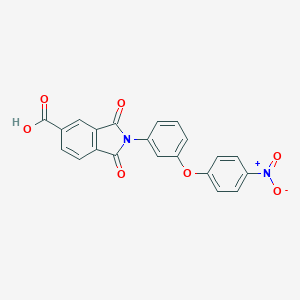 CHEMBL604677 CHEMBL604677 | C21H12N2O7 | 404.334 | 7 / 1 | 3.3 | Yes |
| 114527 | 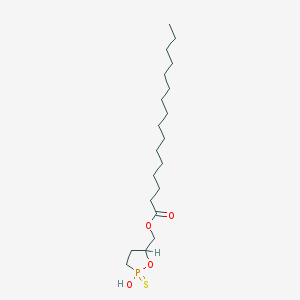 CHEMBL211465 CHEMBL211465 | C20H39O4PS | 406.562 | 5 / 1 | 8.1 | No |
| 117461 | 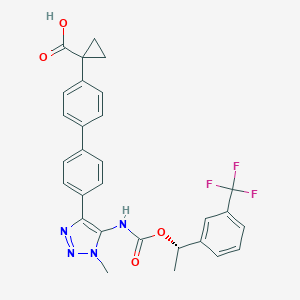 CHEMBL2182047 CHEMBL2182047 | C29H25F3N4O4 | 550.538 | 9 / 2 | 5.5 | No |
| 117462 | 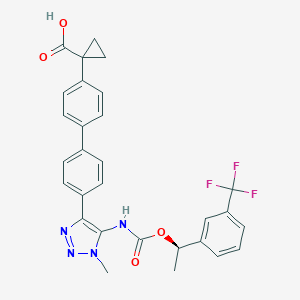 CHEMBL2182046 CHEMBL2182046 | C29H25F3N4O4 | 550.538 | 9 / 2 | 5.5 | No |
| 538874 |  CHEMBL3969018 CHEMBL3969018 | C29H25F3N4O4 | 550.538 | 9 / 2 | 5.5 | No |
| 553866 |  syn-BrP-LPA syn-BrP-LPA | C20H40BrO6P | 487.412 | 6 / 3 | 6.3 | No |
| 553868 |  BrP-LPA BrP-LPA | C20H40BrO6P | 487.412 | 6 / 3 | 6.3 | No |
| 553869 |  anti-BrP-LPA anti-BrP-LPA | C20H40BrO6P | 487.412 | 6 / 3 | 6.3 | No |
| 122947 |  L-NASPA L-NASPA | C19H38NO7P | 423.487 | 7 / 4 | 4.7 | Yes |
| 124787 |  CHEMBL183143 CHEMBL183143 | C35H55N2O7P | 646.806 | 8 / 3 | 8.0 | No |
| 561204 |  SCHEMBL16506802 SCHEMBL16506802 | C30H26FNO5 | 499.538 | 6 / 2 | 5.4 | No |
| 539127 |  CHEMBL3955284 CHEMBL3955284 | C30H26FNO5 | 499.538 | 6 / 2 | 5.4 | No |
| 127641 | 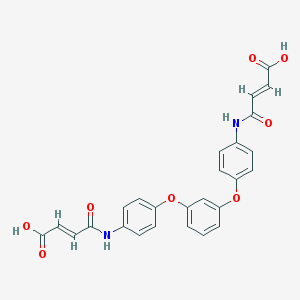 CHEMBL482498 CHEMBL482498 | C26H20N2O8 | 488.452 | 8 / 4 | 2.9 | Yes |
| 127779 | 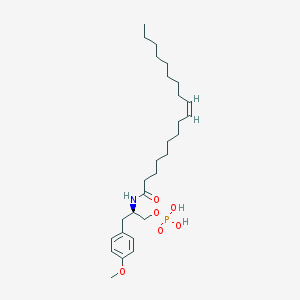 CHEMBL94011 CHEMBL94011 | C28H48NO6P | 525.667 | 6 / 3 | 7.2 | No |
| 129049 | 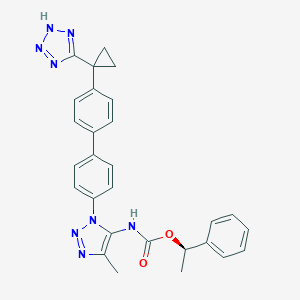 CHEMBL2182038 CHEMBL2182038 | C28H26N8O2 | 506.57 | 7 / 2 | 5.0 | No |
| 130630 | 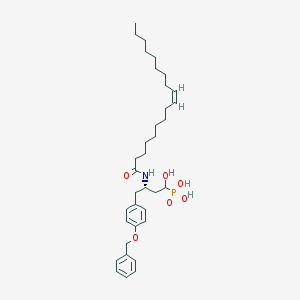 CID 44392848 CID 44392848 | C35H54NO6P | 615.792 | 6 / 4 | 8.5 | No |
| 539299 | 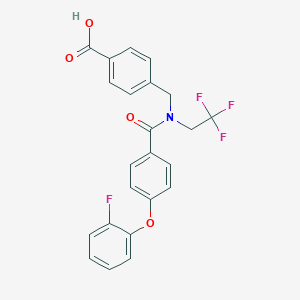 CHEMBL3959098 CHEMBL3959098 | C23H17F4NO4 | 447.386 | 8 / 1 | 5.1 | No |
| 539337 | 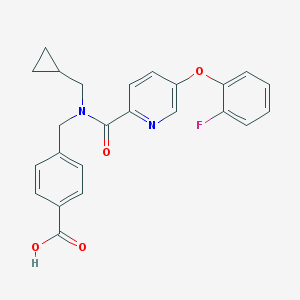 CHEMBL3972525 CHEMBL3972525 | C24H21FN2O4 | 420.44 | 6 / 1 | 4.0 | Yes |
| 135410 | 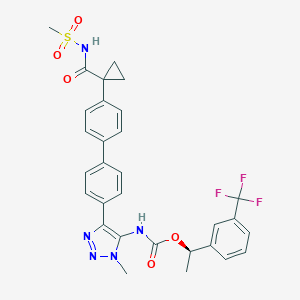 CHEMBL2182050 CHEMBL2182050 | C30H28F3N5O5S | 627.639 | 10 / 2 | 4.8 | No |
| 138416 | 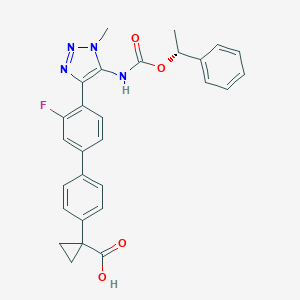 CHEMBL2182044 CHEMBL2182044 | C28H25FN4O4 | 500.53 | 7 / 2 | 4.7 | No |
| 141526 | 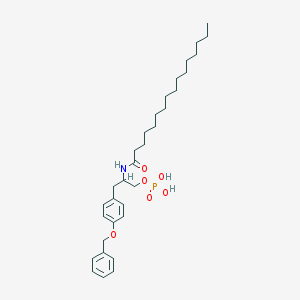 CHEMBL441066 CHEMBL441066 | C32H50NO6P | 575.727 | 6 / 3 | 8.6 | No |
| 141739 | 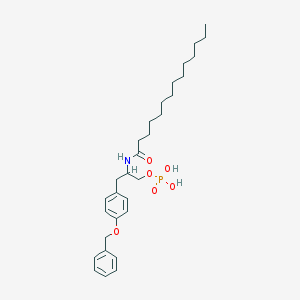 CHEMBL432821 CHEMBL432821 | C30H46NO6P | 547.673 | 6 / 3 | 7.5 | No |
| 144144 | 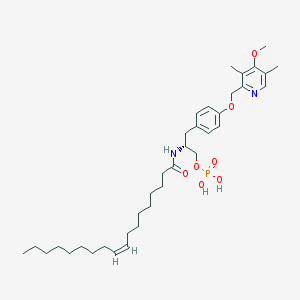 CHEMBL362053 CHEMBL362053 | C36H57N2O7P | 660.833 | 8 / 3 | 8.4 | No |
| 144145 | 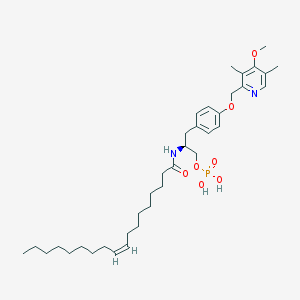 CHEMBL181917 CHEMBL181917 | C36H57N2O7P | 660.833 | 8 / 3 | 8.4 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417