

You can:
| Name | Somatostatin receptor type 1 |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Sstr1 |
| Synonym | SRIF-2 SS-1-R SS1-R SS1R SST1 receptor |
| Disease | N/A for non-human GPCRs |
| Length | 391 |
| Amino acid sequence | MFPNGTAPSPTSSPSSSPGGCGEGVCSRGPGSGAADGMEEPGRNSSQNGTLSEGQGSAILISFIYSVVCLVGLCGNSMVIYVILRYAKMKTATNIYILNLAIADELLMLSVPFLVTSTLLRHWPFGALLCRLVLSVDAVNMFTSIYCLTVLSVDRYVAVVHPIKAARYRRPTVAKVVNLGVWVLSLLVILPIVVFSRTAANSDGTVACNMLMPEPAQRWLVGFVLYTFLMGFLLPVGAICLCYVLIIAKMRMVALKAGWQQRKRSERKITLMVMMVVMVFVICWMPFYVVQLVNVFAEQDDATVSQLSVILGYANSCANPILYGFLSDNFKRSFQRILCLSWMDNAAEEPVDYYATALKSRAYSVEDFQPENLESGGVFRNGTCASRISTL |
| UniProt | P28646 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL4652 |
| IUPHAR | 355 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 856 | 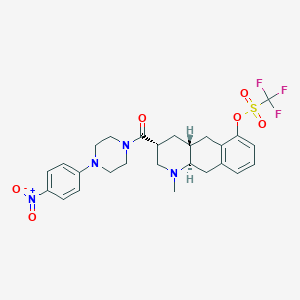 CHEMBL399843 CHEMBL399843 | C26H29F3N4O6S | 582.595 | 11 / 0 | 4.3 | No |
| 5068 | 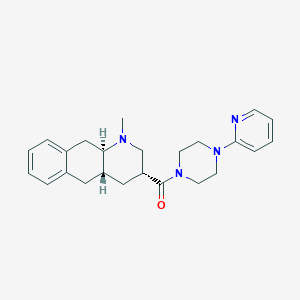 CHEMBL251335 CHEMBL251335 | C24H30N4O | 390.531 | 4 / 0 | 3.0 | Yes |
| 5506 | 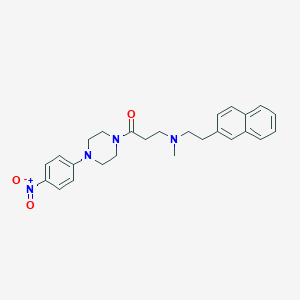 CHEMBL460934 CHEMBL460934 | C26H30N4O3 | 446.551 | 5 / 0 | 4.3 | Yes |
| 6804 | 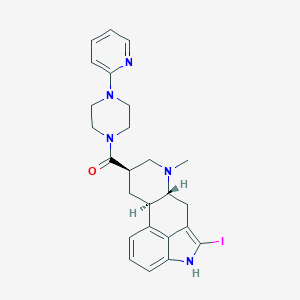 CHEMBL253653 CHEMBL253653 | C25H28IN5O | 541.437 | 4 / 1 | 3.4 | No |
| 11126 | 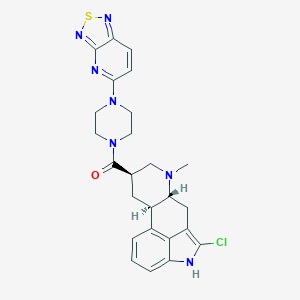 CHEMBL252032 CHEMBL252032 | C25H26ClN7OS | 508.041 | 7 / 1 | 3.9 | No |
| 12116 | 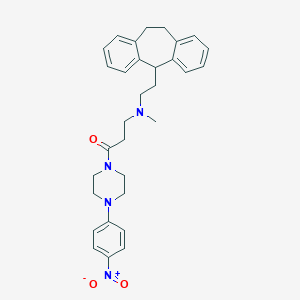 CHEMBL516837 CHEMBL516837 | C31H36N4O3 | 512.654 | 5 / 0 | 5.2 | No |
| 12320 | 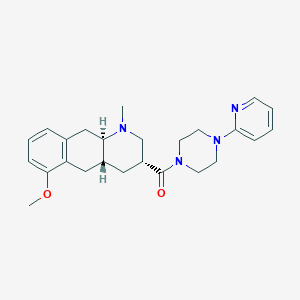 CHEMBL251334 CHEMBL251334 | C25H32N4O2 | 420.557 | 5 / 0 | 3.0 | Yes |
| 15955 | 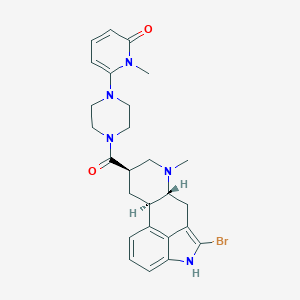 CHEMBL254500 CHEMBL254500 | C26H30BrN5O2 | 524.463 | 4 / 1 | 3.1 | No |
| 15966 | 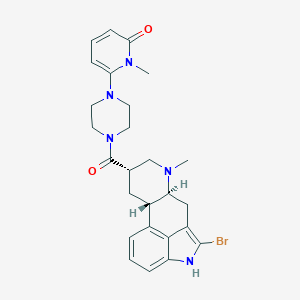 CHEMBL252231 CHEMBL252231 | C26H30BrN5O2 | 524.463 | 4 / 1 | 3.1 | No |
| 16294 | 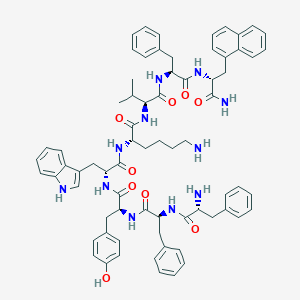 BDBM84629 BDBM84629 | C71H81N11O9 | 1232.5 | 11 / 12 | 8.1 | No |
| 22097 | 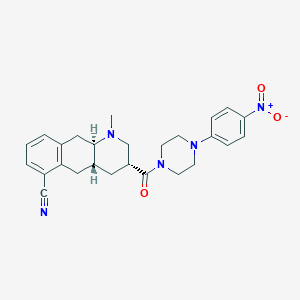 CHEMBL401091 CHEMBL401091 | C26H29N5O3 | 459.55 | 6 / 0 | 3.3 | Yes |
| 22965 | 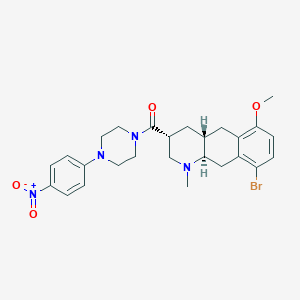 CHEMBL248716 CHEMBL248716 | C26H31BrN4O4 | 543.462 | 6 / 0 | 4.3 | No |
| 24863 | 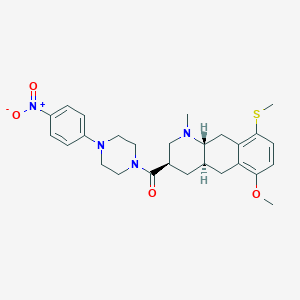 CHEMBL248717 CHEMBL248717 | C27H34N4O4S | 510.653 | 7 / 0 | 4.1 | No |
| 28102 | 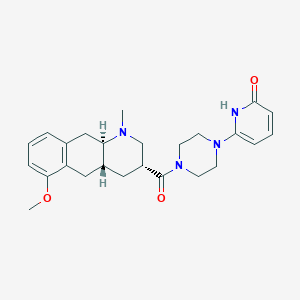 CHEMBL398655 CHEMBL398655 | C25H32N4O3 | 436.556 | 5 / 1 | 2.2 | Yes |
| 31477 | 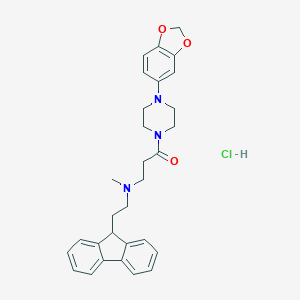 CHEMBL491498 CHEMBL491498 | C30H34ClN3O3 | 520.07 | 5 / 1 | N/A | No |
| 32621 | 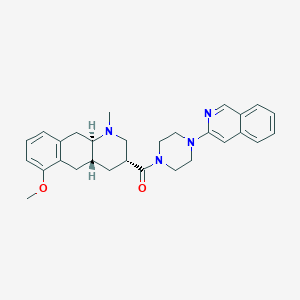 CHEMBL440962 CHEMBL440962 | C29H34N4O2 | 470.617 | 5 / 0 | 4.3 | Yes |
| 35386 | 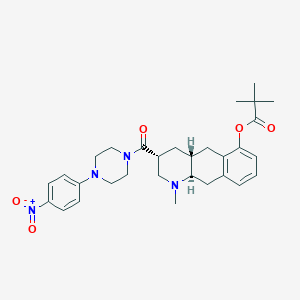 CHEMBL251529 CHEMBL251529 | C30H38N4O5 | 534.657 | 7 / 0 | 4.7 | No |
| 40413 | 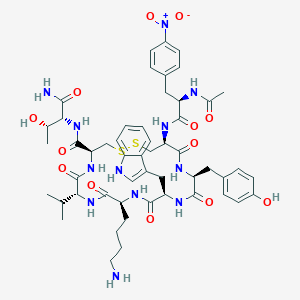 BDBM85012 BDBM85012 | C52H68N12O13S2 | 1133.31 | 16 / 13 | 1.3 | No |
| 40768 | 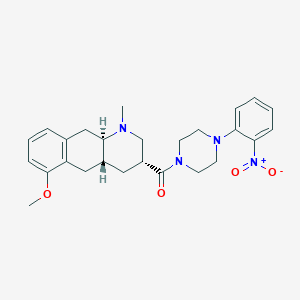 CHEMBL398681 CHEMBL398681 | C26H32N4O4 | 464.566 | 6 / 0 | 3.6 | Yes |
| 42101 | 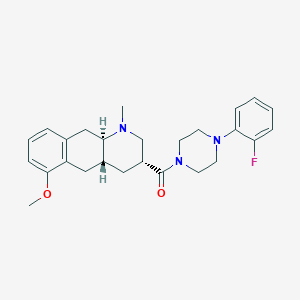 CHEMBL251311 CHEMBL251311 | C26H32FN3O2 | 437.559 | 5 / 0 | 3.8 | Yes |
| 46311 | 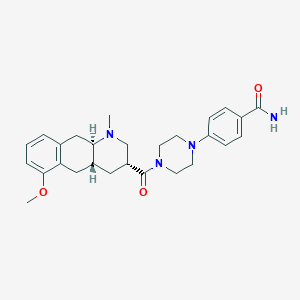 CHEMBL249892 CHEMBL249892 | C27H34N4O3 | 462.594 | 5 / 1 | 2.6 | Yes |
| 48332 | 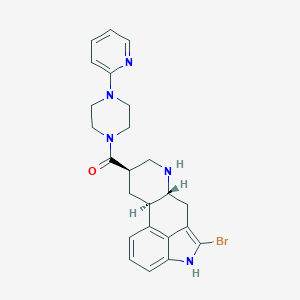 CHEMBL252002 CHEMBL252002 | C24H26BrN5O | 480.41 | 4 / 2 | 3.2 | Yes |
| 51728 | 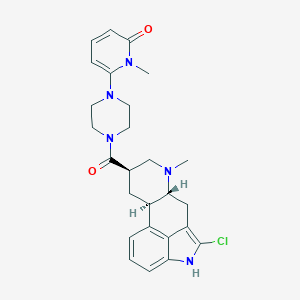 CHEMBL254499 CHEMBL254499 | C26H30ClN5O2 | 480.009 | 4 / 1 | 3.1 | Yes |
| 54300 | 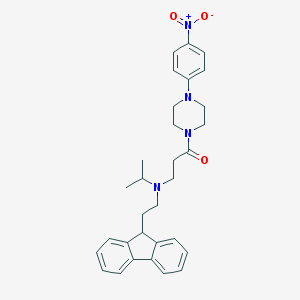 CHEMBL514027 CHEMBL514027 | C31H36N4O3 | 512.654 | 5 / 0 | 5.4 | No |
| 54958 | 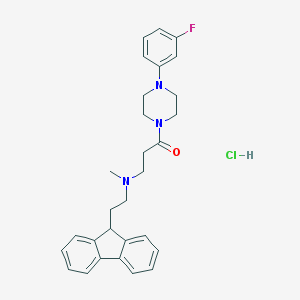 CHEMBL472752 CHEMBL472752 | C29H33ClFN3O | 494.051 | 4 / 1 | N/A | N/A |
| 56742 | 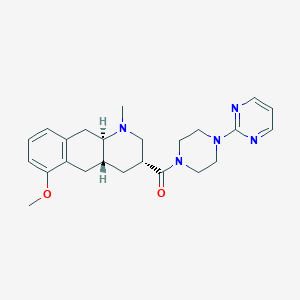 CHEMBL400677 CHEMBL400677 | C24H31N5O2 | 421.545 | 6 / 0 | 2.4 | Yes |
| 62259 | 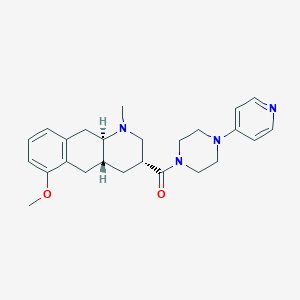 CHEMBL250111 CHEMBL250111 | C25H32N4O2 | 420.557 | 5 / 0 | 2.7 | Yes |
| 63618 | 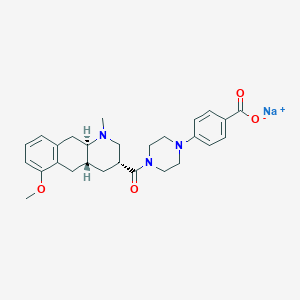 CHEMBL249894 CHEMBL249894 | C27H32N3NaO4 | 485.56 | 6 / 0 | N/A | N/A |
| 68007 | 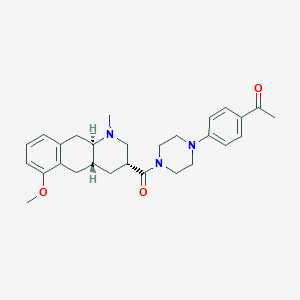 CHEMBL250705 CHEMBL250705 | C28H35N3O3 | 461.606 | 5 / 0 | 3.4 | Yes |
| 70044 | 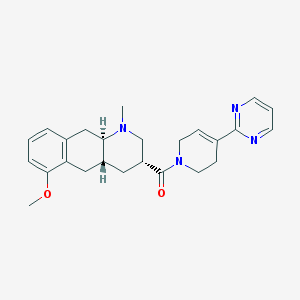 CHEMBL251110 CHEMBL251110 | C25H30N4O2 | 418.541 | 5 / 0 | 2.3 | Yes |
| 71498 | 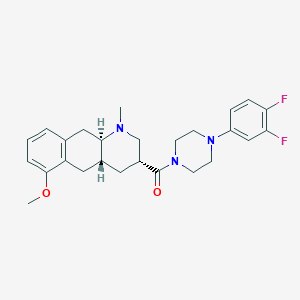 CHEMBL250304 CHEMBL250304 | C26H31F2N3O2 | 455.55 | 6 / 0 | 3.9 | Yes |
| 73234 | 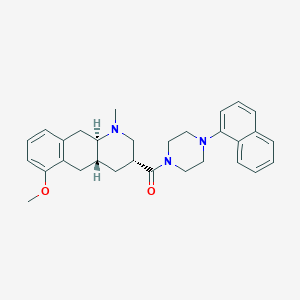 CHEMBL250521 CHEMBL250521 | C30H35N3O2 | 469.629 | 4 / 0 | 5.0 | Yes |
| 74290 | 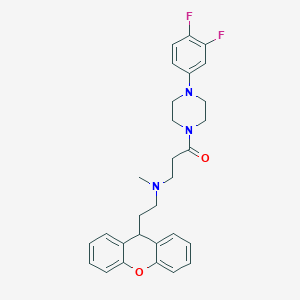 CHEMBL460542 CHEMBL460542 | C29H31F2N3O2 | 491.583 | 6 / 0 | 4.9 | Yes |
| 75961 | 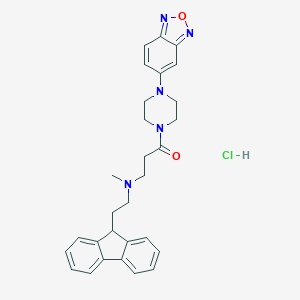 CHEMBL523814 CHEMBL523814 | C29H32ClN5O2 | 518.058 | 6 / 1 | N/A | No |
| 80046 | 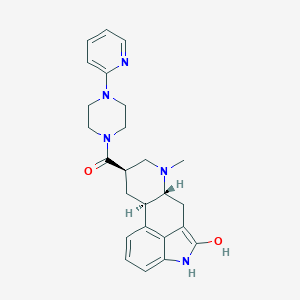 CHEMBL253654 CHEMBL253654 | C25H29N5O2 | 431.54 | 5 / 2 | 2.7 | Yes |
| 89004 | 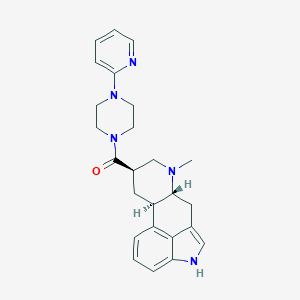 CHEMBL253232 CHEMBL253232 | C25H29N5O | 415.541 | 4 / 1 | 2.7 | Yes |
| 89450 | 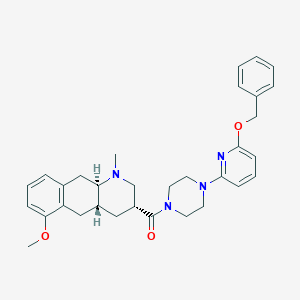 CHEMBL251116 CHEMBL251116 | C32H38N4O3 | 526.681 | 6 / 0 | 4.8 | No |
| 92137 | 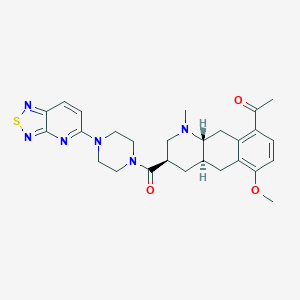 CHEMBL249111 CHEMBL249111 | C27H32N6O3S | 520.652 | 9 / 0 | 2.9 | No |
| 99985 | 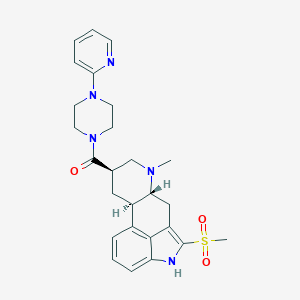 CHEMBL401108 CHEMBL401108 | C26H31N5O3S | 493.626 | 6 / 1 | 2.3 | Yes |
| 101077 | 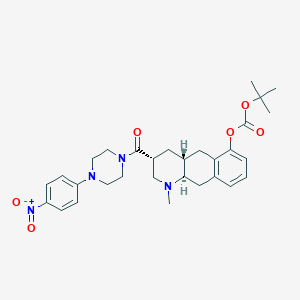 CHEMBL251751 CHEMBL251751 | C30H38N4O6 | 550.656 | 8 / 0 | 4.7 | No |
| 101903 | 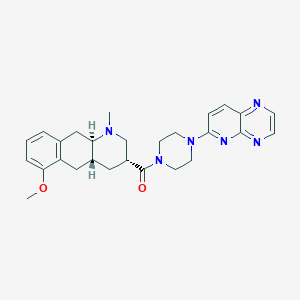 CHEMBL398656 CHEMBL398656 | C27H32N6O2 | 472.593 | 7 / 0 | 2.6 | Yes |
| 103288 | 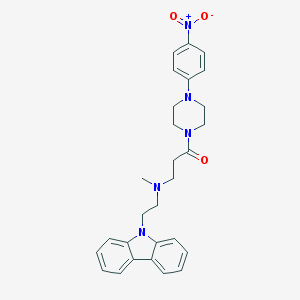 CHEMBL462157 CHEMBL462157 | C28H31N5O3 | 485.588 | 5 / 0 | 4.2 | Yes |
| 104523 | 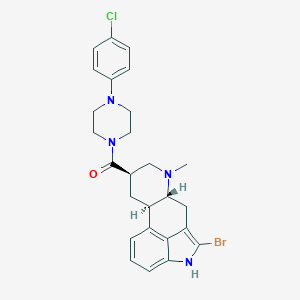 CHEMBL539415 CHEMBL539415 | C26H28BrClN4O | 527.891 | 3 / 1 | 5.1 | No |
| 106158 | 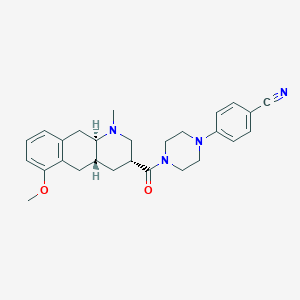 CHEMBL249932 CHEMBL249932 | C27H32N4O2 | 444.579 | 5 / 0 | 3.5 | Yes |
| 108135 | 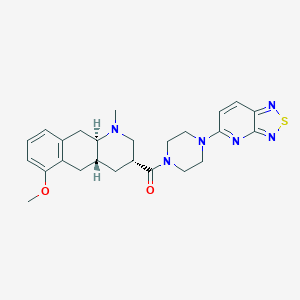 CHEMBL248715 CHEMBL248715 | C25H30N6O2S | 478.615 | 8 / 0 | 3.2 | Yes |
| 108830 | 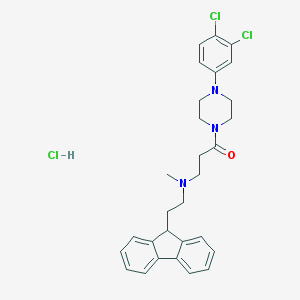 CHEMBL472751 CHEMBL472751 | C29H32Cl3N3O | 544.945 | 3 / 1 | N/A | No |
| 108956 | 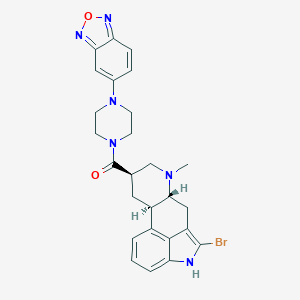 CHEMBL251835 CHEMBL251835 | C26H27BrN6O2 | 535.446 | 6 / 1 | 3.7 | No |
| 108964 | 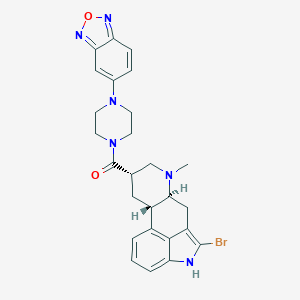 CHEMBL252232 CHEMBL252232 | C26H27BrN6O2 | 535.446 | 6 / 1 | 3.7 | No |
| 109780 | 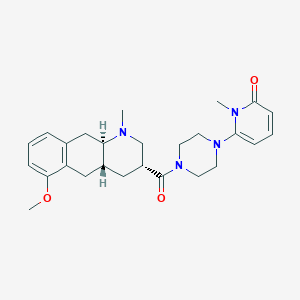 CHEMBL440235 CHEMBL440235 | C26H34N4O3 | 450.583 | 5 / 0 | 2.4 | Yes |
| 112066 | 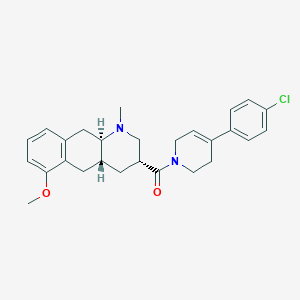 CHEMBL250912 CHEMBL250912 | C27H31ClN2O2 | 451.007 | 3 / 0 | 4.6 | Yes |
| 115896 | 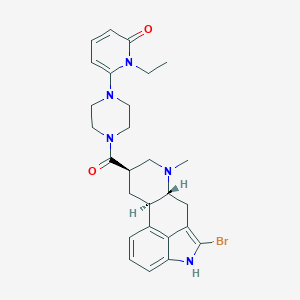 CHEMBL254505 CHEMBL254505 | C27H32BrN5O2 | 538.49 | 4 / 1 | 3.5 | No |
| 117218 | 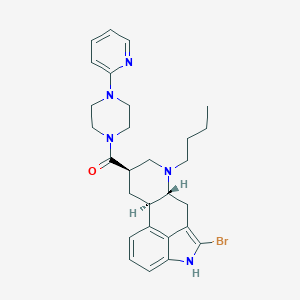 CHEMBL445673 CHEMBL445673 | C28H34BrN5O | 536.518 | 4 / 1 | 5.0 | No |
| 120623 | 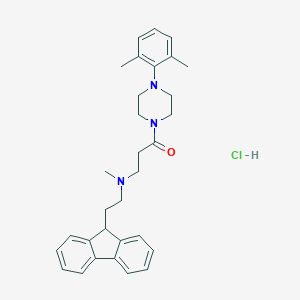 CHEMBL491143 CHEMBL491143 | C31H38ClN3O | 504.115 | 3 / 1 | N/A | No |
| 123172 | 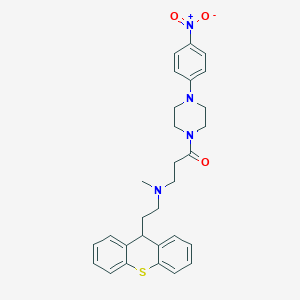 CHEMBL462335 CHEMBL462335 | C29H32N4O3S | 516.66 | 6 / 0 | 5.0 | No |
| 124833 | 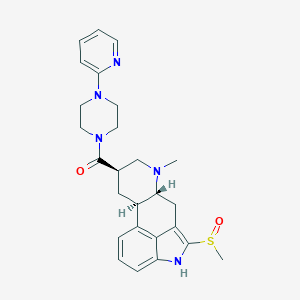 CHEMBL251801 CHEMBL251801 | C26H31N5O2S | 477.627 | 6 / 1 | 2.1 | Yes |
| 126612 | 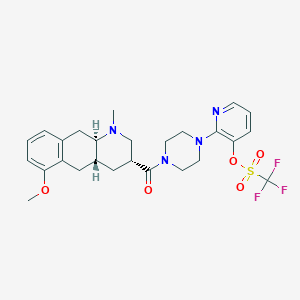 CHEMBL250314 CHEMBL250314 | C26H31F3N4O5S | 568.612 | 11 / 0 | 3.8 | No |
| 127322 | 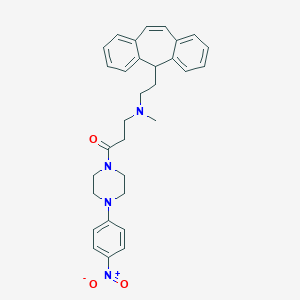 CHEMBL517921 CHEMBL517921 | C31H34N4O3 | 510.638 | 5 / 0 | 5.4 | No |
| 127719 | 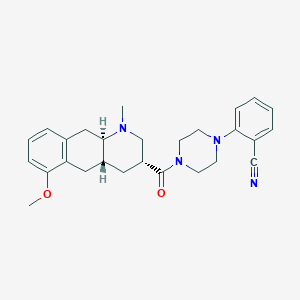 CHEMBL249930 CHEMBL249930 | C27H32N4O2 | 444.579 | 5 / 0 | 3.5 | Yes |
| 128571 | 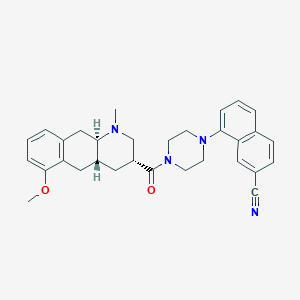 CHEMBL250522 CHEMBL250522 | C31H34N4O2 | 494.639 | 5 / 0 | 4.7 | Yes |
| 131134 | 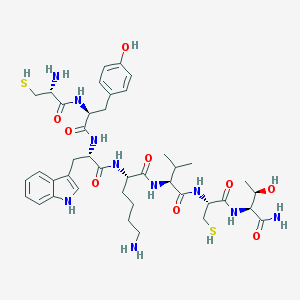 BDBM82254 BDBM82254 | C41H60N10O9S2 | 901.112 | 13 / 14 | 0.0 | No |
| 133228 | 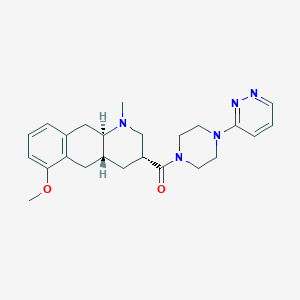 CHEMBL400481 CHEMBL400481 | C24H31N5O2 | 421.545 | 6 / 0 | 2.0 | Yes |
| 134607 | 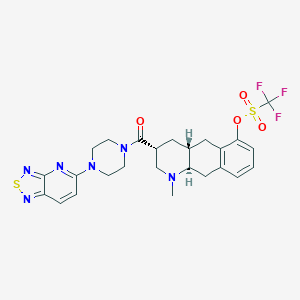 CHEMBL251754 CHEMBL251754 | C25H27F3N6O4S2 | 596.644 | 13 / 0 | 4.0 | No |
| 135040 | 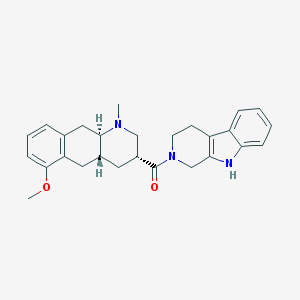 CHEMBL251300 CHEMBL251300 | C27H31N3O2 | 429.564 | 3 / 1 | 3.8 | Yes |
| 135047 | 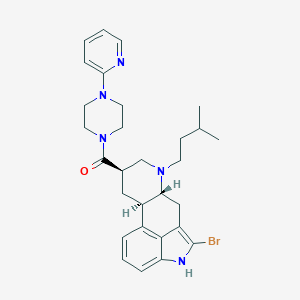 CHEMBL253880 CHEMBL253880 | C29H36BrN5O | 550.545 | 4 / 1 | 5.4 | No |
| 137631 | 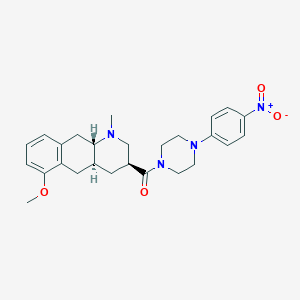 CHEMBL249112 CHEMBL249112 | C26H32N4O4 | 464.566 | 6 / 0 | 3.6 | Yes |
| 137636 | 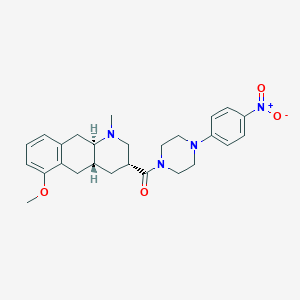 Obelin Obelin | C26H32N4O4 | 464.566 | 6 / 0 | 3.6 | Yes |
| 138471 | 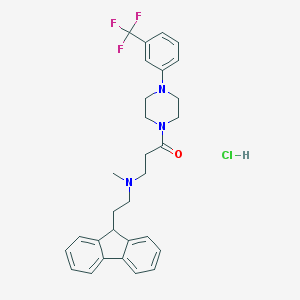 CHEMBL495264 CHEMBL495264 | C30H33ClF3N3O | 544.059 | 6 / 1 | N/A | No |
| 142207 | 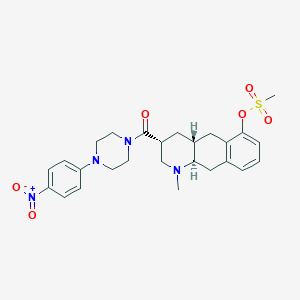 CHEMBL440236 CHEMBL440236 | C26H32N4O6S | 528.624 | 8 / 0 | 3.1 | No |
| 142937 | 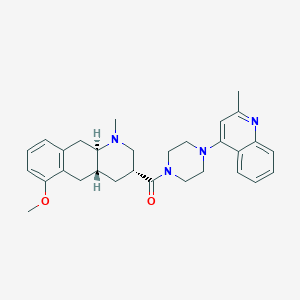 CHEMBL250723 CHEMBL250723 | C30H36N4O2 | 484.644 | 5 / 0 | 4.4 | Yes |
| 144428 | 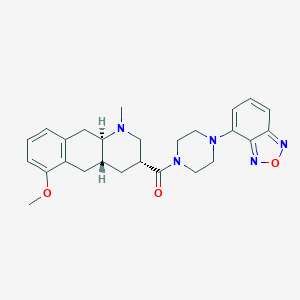 CHEMBL250921 CHEMBL250921 | C26H31N5O3 | 461.566 | 7 / 0 | 3.0 | Yes |
| 145345 | 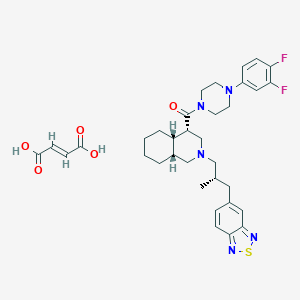 CHEMBL1076623 CHEMBL1076623 | C34H41F2N5O5S | 669.789 | 12 / 2 | N/A | No |
| 145491 | 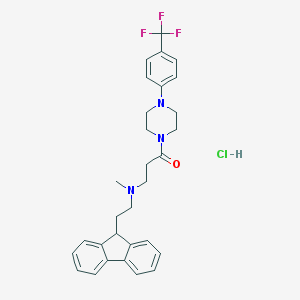 CHEMBL470500 CHEMBL470500 | C30H33ClF3N3O | 544.059 | 6 / 1 | N/A | No |
| 155584 | 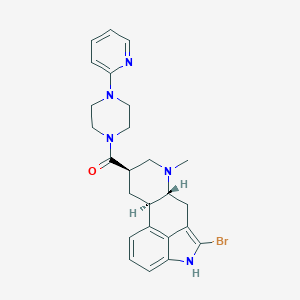 CHEMBL253438 CHEMBL253438 | C25H28BrN5O | 494.437 | 4 / 1 | 3.7 | Yes |
| 158431 | 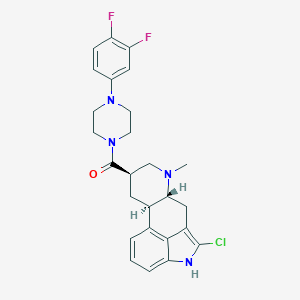 CHEMBL252428 CHEMBL252428 | C26H27ClF2N4O | 484.976 | 5 / 1 | 4.6 | Yes |
| 163377 | 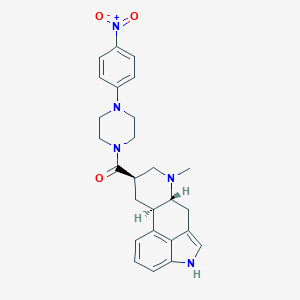 CHEMBL252029 CHEMBL252029 | C26H29N5O3 | 459.55 | 5 / 1 | 3.3 | Yes |
| 163791 | 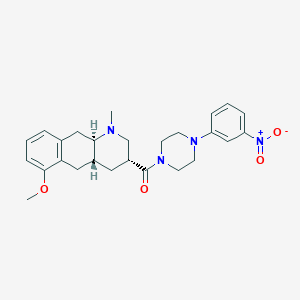 CHEMBL250498 CHEMBL250498 | C26H32N4O4 | 464.566 | 6 / 0 | 3.6 | Yes |
| 166603 | 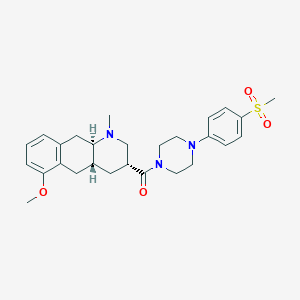 CHEMBL250098 CHEMBL250098 | C27H35N3O4S | 497.654 | 6 / 0 | 3.0 | Yes |
| 170584 | 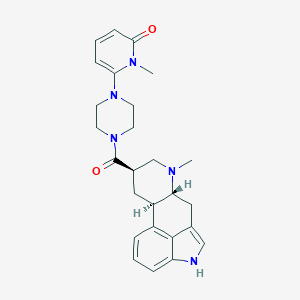 CHEMBL254288 CHEMBL254288 | C26H31N5O2 | 445.567 | 4 / 1 | 2.1 | Yes |
| 175221 | 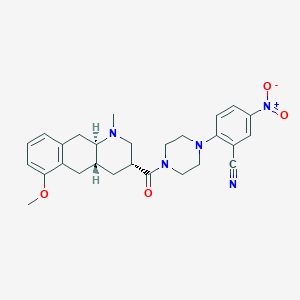 CHEMBL250305 CHEMBL250305 | C27H31N5O4 | 489.576 | 7 / 0 | 3.3 | Yes |
| 175936 | 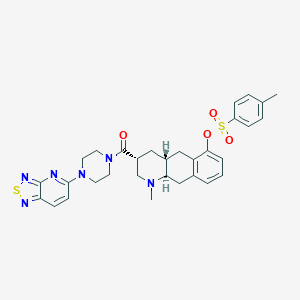 CHEMBL251753 CHEMBL251753 | C31H34N6O4S2 | 618.771 | 10 / 0 | 4.7 | No |
| 177225 | 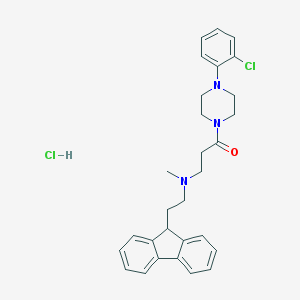 CHEMBL472750 CHEMBL472750 | C29H33Cl2N3O | 510.503 | 3 / 1 | N/A | No |
| 186583 | 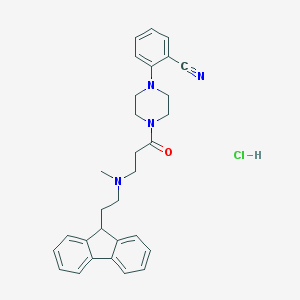 CHEMBL491508 CHEMBL491508 | C30H33ClN4O | 501.071 | 4 / 1 | N/A | No |
| 195431 | 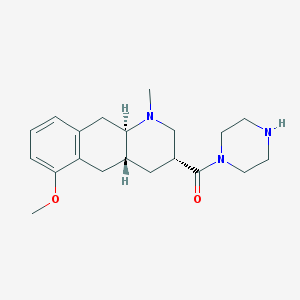 CHEMBL251301 CHEMBL251301 | C20H29N3O2 | 343.471 | 4 / 1 | 1.5 | Yes |
| 195865 | 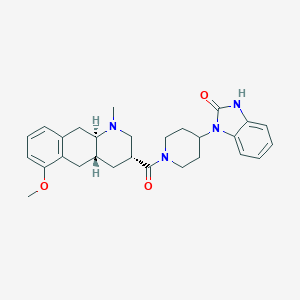 CHEMBL251111 CHEMBL251111 | C28H34N4O3 | 474.605 | 4 / 1 | 3.2 | Yes |
| 197344 | 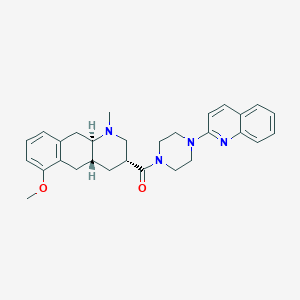 CHEMBL250724 CHEMBL250724 | C29H34N4O2 | 470.617 | 5 / 0 | 4.3 | Yes |
| 202250 | 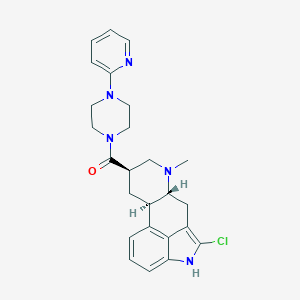 CHEMBL253234 CHEMBL253234 | C25H28ClN5O | 449.983 | 4 / 1 | 3.7 | Yes |
| 206828 | 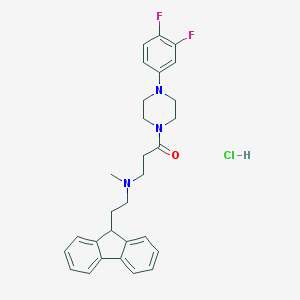 CHEMBL473763 CHEMBL473763 | C29H32ClF2N3O | 512.042 | 5 / 1 | N/A | No |
| 210154 | 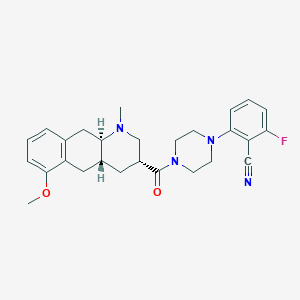 CHEMBL400136 CHEMBL400136 | C27H31FN4O2 | 462.569 | 6 / 0 | 3.6 | Yes |
| 210207 | 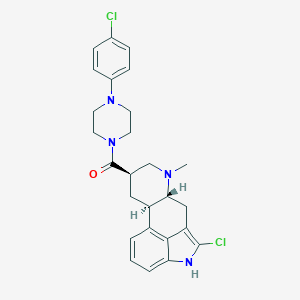 CHEMBL401131 CHEMBL401131 | C26H28Cl2N4O | 483.437 | 3 / 1 | 5.0 | Yes |
| 211596 | 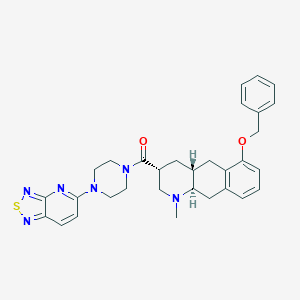 CHEMBL399464 CHEMBL399464 | C31H34N6O2S | 554.713 | 8 / 0 | 4.7 | No |
| 213988 | 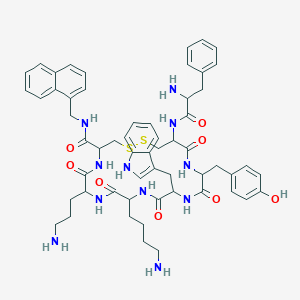 BDBM82255 BDBM82255 | C57H69N11O8S2 | 1100.37 | 13 / 12 | 3.7 | No |
| 215262 | 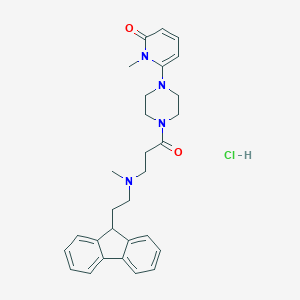 CHEMBL491144 CHEMBL491144 | C29H35ClN4O2 | 507.075 | 4 / 1 | N/A | No |
| 215819 | 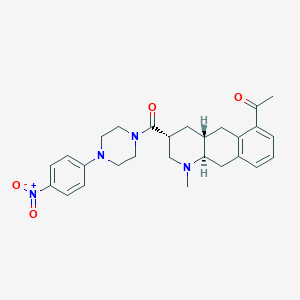 CHEMBL251752 CHEMBL251752 | C27H32N4O4 | 476.577 | 6 / 0 | 3.3 | Yes |
| 219097 | 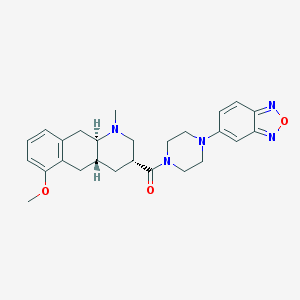 CHEMBL250922 CHEMBL250922 | C26H31N5O3 | 461.566 | 7 / 0 | 3.0 | Yes |
| 219837 | 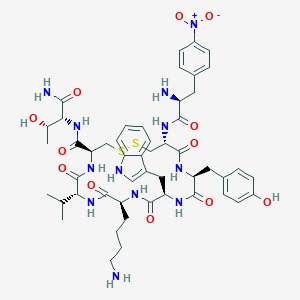 BDBM85009 BDBM85009 | C50H66N12O12S2 | 1091.27 | 16 / 13 | 1.1 | No |
| 219841 | 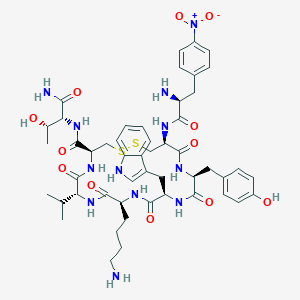 BDBM85013 BDBM85013 | C50H66N12O12S2 | 1091.27 | 16 / 13 | 1.1 | No |
| 220116 | 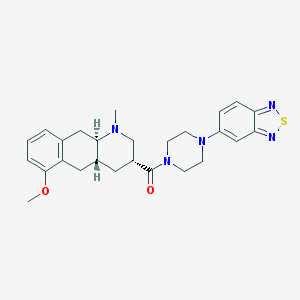 CHEMBL400099 CHEMBL400099 | C26H31N5O2S | 477.627 | 7 / 0 | 3.6 | Yes |
| 223715 | 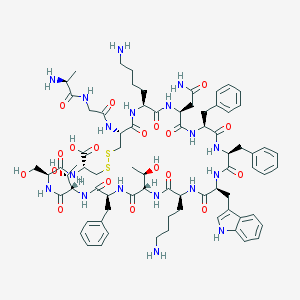 SOMATOSTATIN SOMATOSTATIN | C76H104N18O19S2 | 1637.9 | 24 / 22 | -3.1 | No |
| 223720 | 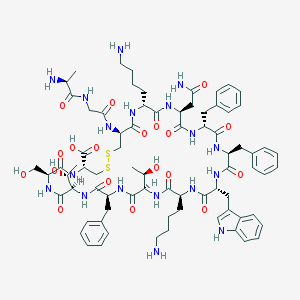 Somatostatin-14 Somatostatin-14 | C76H104N18O19S2 | 1637.9 | 24 / 22 | -3.1 | No |
| 226328 | 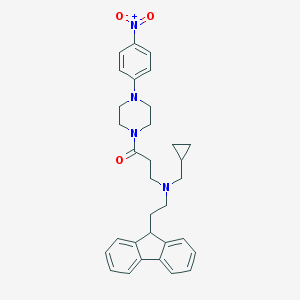 CHEMBL470331 CHEMBL470331 | C32H36N4O3 | 524.665 | 5 / 0 | 5.3 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417