

You can:
| Name | Metabotropic glutamate receptor 2 |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Grm2 |
| Synonym | glutamate receptor GPRC1B metabotropic glutamate receptor 2 mGlu2 receptor mGluR2 |
| Disease | N/A for non-human GPCRs |
| Length | 872 |
| Amino acid sequence | MESLLGFLALLLLWGAVAEGPAKKVLTLEGDLVLGGLFPVHQKGGPAEECGPVNEHRGIQRLEAMLFALDRINRDPHLLPGVRLGAHILDSCSKDTHALEQALDFVRASLSRGADGSRHICPDGSYATHSDAPTAVTGVIGGSYSDVSIQVANLLRLFQIPQISYASTSAKLSDKSRYDYFARTVPPDFFQAKAMAEILRFFNWTYVSTVASEGDYGETGIEAFELEARARNICVATSEKVGRAMSRAAFEGVVRALLQKPSARVAVLFTRSEDARELLAATQRLNASFTWVASDGWGALESVVAGSERAAEGAITIELASYPISDFASYFQSLDPWNNSRNPWFREFWEERFHCSFRQRDCAAHSLRAVPFEQESKIMFVVNAVYAMAHALHNMHRALCPNTTHLCDAMRPVNGRRLYKDFVLNVKFDAPFRPADTDDEVRFDRFGDGIGRYNIFTYLRAGSGRYRYQKVGYWAEGLTLDTSFIPWASPSAGPLPASRCSEPCLQNEVKSVQPGEVCCWLCIPCQPYEYRLDEFTCADCGLGYWPNASLTGCFELPQEYIRWGDAWAVGPVTIACLGALATLFVLGVFVRHNATPVVKASGRELCYILLGGVFLCYCMTFVFIAKPSTAVCTLRRLGLGTAFSVCYSALLTKTNRIARIFGGAREGAQRPRFISPASQVAICLALISGQLLIVAAWLVVEAPGTGKETAPERREVVTLRCNHRDASMLGSLAYNVLLIALCTLYAFKTRKCPENFNEAKFIGFTMYTTCIIWLAFLPIFYVTSSDYRVQTTTMCVSVSLSGSVVLGCLFAPKLHIILFQPQKNVVSHRAPTSRFGSAAPRASANLGQGSGSQFVPTVCNGREVVDSTTSSL |
| UniProt | P31421 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL2851 |
| IUPHAR | 290 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 110 | 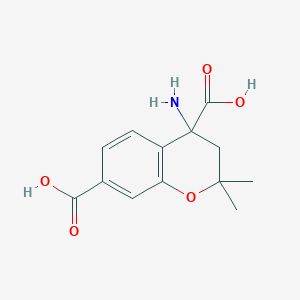 CHEMBL278949 CHEMBL278949 | C13H15NO5 | 265.265 | 6 / 3 | -1.6 | Yes |
| 230 | 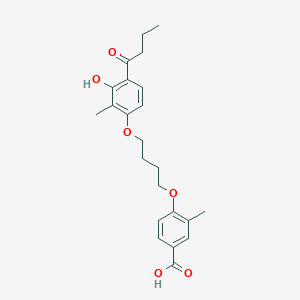 CHEMBL3287722 CHEMBL3287722 | C23H28O6 | 400.471 | 6 / 2 | 5.2 | No |
| 938 | 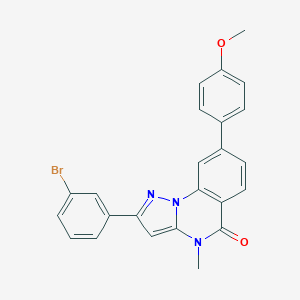 CHEMBL3288658 CHEMBL3288658 | C24H18BrN3O2 | 460.331 | 3 / 0 | 5.3 | No |
| 1227 | 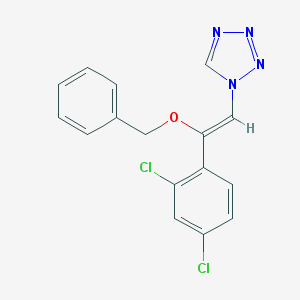 CHEMBL69204 CHEMBL69204 | C16H12Cl2N4O | 347.199 | 4 / 0 | 4.2 | Yes |
| 1358 | 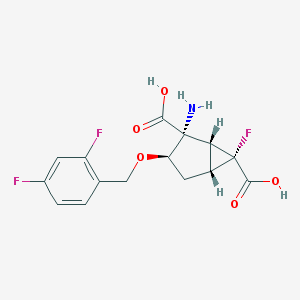 CHEMBL188511 CHEMBL188511 | C15H14F3NO5 | 345.274 | 9 / 3 | -1.7 | Yes |
| 4431 | 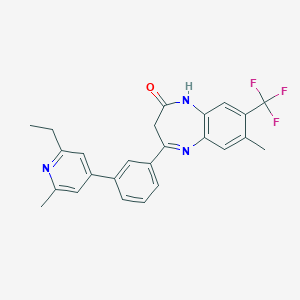 CHEMBL1629856 CHEMBL1629856 | C25H22F3N3O | 437.466 | 6 / 1 | 5.5 | No |
| 5897 | 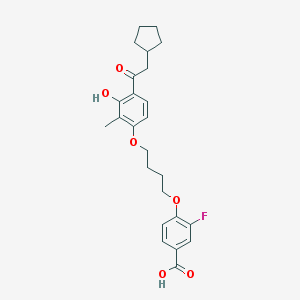 CHEMBL3287694 CHEMBL3287694 | C25H29FO6 | 444.499 | 7 / 2 | 6.3 | No |
| 6650 | 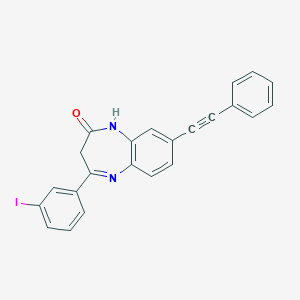 CHEMBL236035 CHEMBL236035 | C23H15IN2O | 462.29 | 2 / 1 | 5.1 | No |
| 6713 | 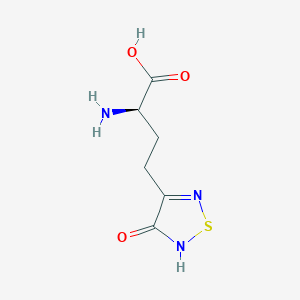 CHEMBL334842 CHEMBL334842 | C6H9N3O3S | 203.216 | 6 / 3 | -3.4 | Yes |
| 6716 | 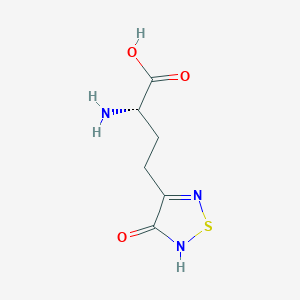 CHEMBL131922 CHEMBL131922 | C6H9N3O3S | 203.216 | 6 / 3 | -3.4 | Yes |
| 7558 | 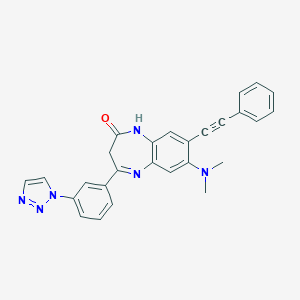 CHEMBL270402 CHEMBL270402 | C27H22N6O | 446.514 | 5 / 1 | 4.0 | Yes |
| 8692 | 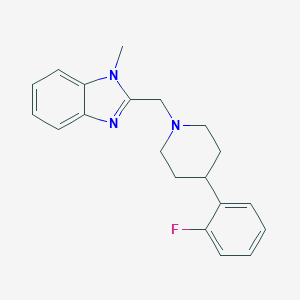 CHEMBL607689 CHEMBL607689 | C20H22FN3 | 323.415 | 3 / 0 | 3.6 | Yes |
| 9259 |  CHEMBL2179623 CHEMBL2179623 | C24H17ClNNaO4S | 473.903 | 5 / 0 | N/A | N/A |
| 459301 |  CHEMBL543097 CHEMBL543097 | C14H18N2O5 | 294.307 | 7 / 3 | -4.6 | Yes |
| 10715 |  CHEMBL3287723 CHEMBL3287723 | C25H32O7 | 444.524 | 7 / 2 | 5.7 | No |
| 521771 |  CHEMBL2312685 CHEMBL2312685 | C8H16N2O4 | 204.226 | 6 / 4 | -6.8 | Yes |
| 11663 | 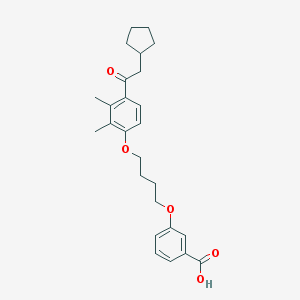 CHEMBL3287674 CHEMBL3287674 | C26H32O5 | 424.537 | 5 / 1 | 6.3 | No |
| 12255 | 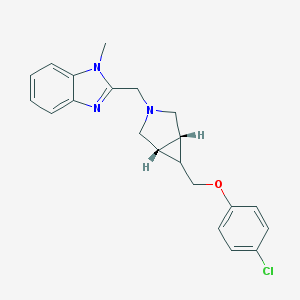 CHEMBL477170 CHEMBL477170 | C21H22ClN3O | 367.877 | 3 / 0 | 3.9 | Yes |
| 12909 | 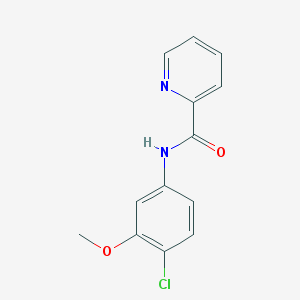 VU 0361737 VU 0361737 | C13H11ClN2O2 | 262.693 | 3 / 1 | 2.6 | Yes |
| 13314 | 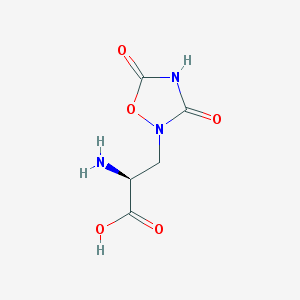 QUISQUALIC ACID QUISQUALIC ACID | C5H7N3O5 | 189.127 | 6 / 3 | -3.9 | Yes |
| 15042 | 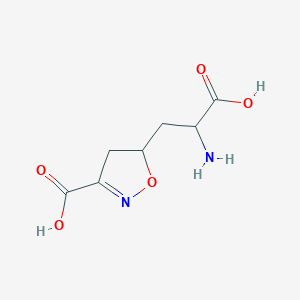 CHEMBL197629 CHEMBL197629 | C7H10N2O5 | 202.166 | 7 / 3 | -3.4 | Yes |
| 16026 | 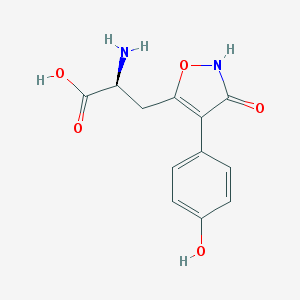 CHEMBL367027 CHEMBL367027 | C12H12N2O5 | 264.237 | 6 / 4 | -2.3 | Yes |
| 16562 | 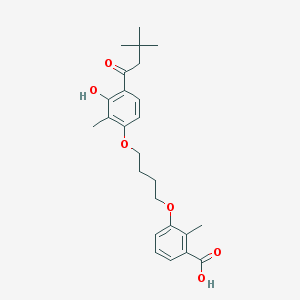 CHEMBL3287725 CHEMBL3287725 | C25H32O6 | 428.525 | 6 / 2 | 6.1 | No |
| 17555 | 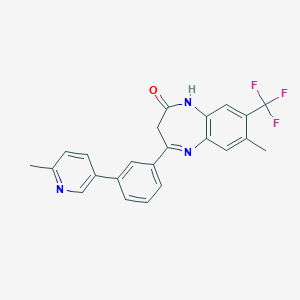 CHEMBL1629845 CHEMBL1629845 | C23H18F3N3O | 409.412 | 6 / 1 | 4.6 | Yes |
| 18148 | 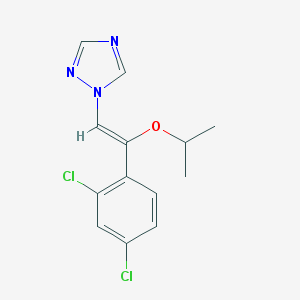 CHEMBL304285 CHEMBL304285 | C13H13Cl2N3O | 298.167 | 3 / 0 | 3.9 | Yes |
| 18628 | 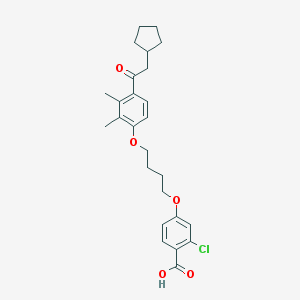 CHEMBL3287682 CHEMBL3287682 | C26H31ClO5 | 458.979 | 5 / 1 | 7.0 | No |
| 19342 | 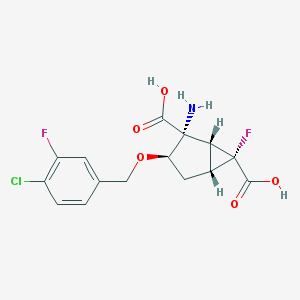 CHEMBL362325 CHEMBL362325 | C15H14ClF2NO5 | 361.726 | 8 / 3 | -1.2 | Yes |
| 19407 | 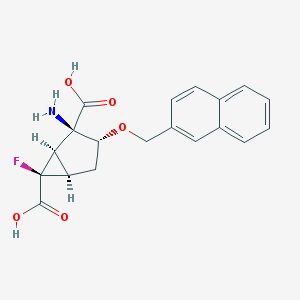 CHEMBL185210 CHEMBL185210 | C19H18FNO5 | 359.353 | 7 / 3 | -0.7 | Yes |
| 20314 | 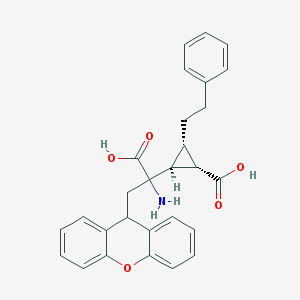 CHEMBL319279 CHEMBL319279 | C28H27NO5 | 457.526 | 6 / 3 | 2.1 | Yes |
| 21776 | 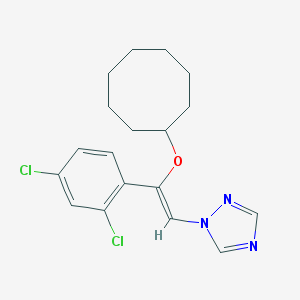 CHEMBL68739 CHEMBL68739 | C18H21Cl2N3O | 366.286 | 3 / 0 | 6.0 | No |
| 21930 | 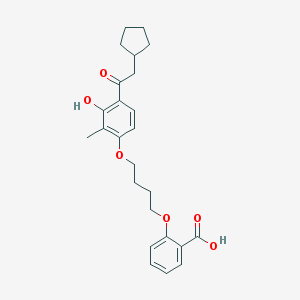 CHEMBL3287680 CHEMBL3287680 | C25H30O6 | 426.509 | 6 / 2 | 6.2 | No |
| 22394 | 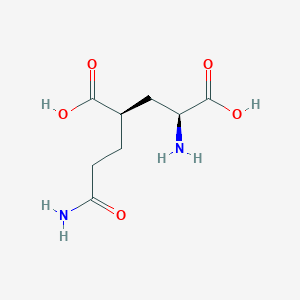 CHEMBL479804 CHEMBL479804 | C8H14N2O5 | 218.209 | 6 / 4 | -3.3 | Yes |
| 22707 | 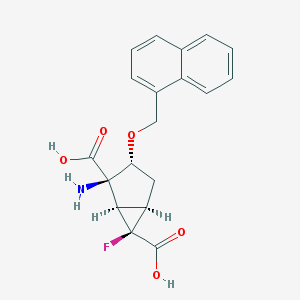 CHEMBL364201 CHEMBL364201 | C19H18FNO5 | 359.353 | 7 / 3 | -0.7 | Yes |
| 23807 | 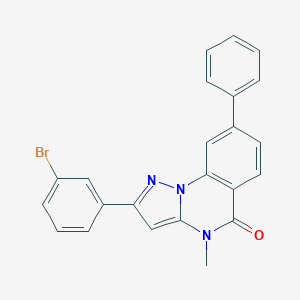 CHEMBL3288657 CHEMBL3288657 | C23H16BrN3O | 430.305 | 2 / 0 | 5.3 | No |
| 522259 | 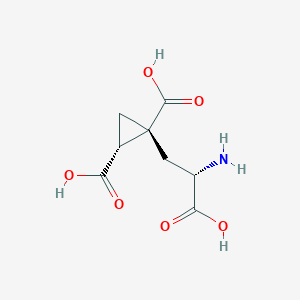 CHEMBL3786667 CHEMBL3786667 | C8H11NO6 | 217.177 | 7 / 4 | -4.0 | Yes |
| 522270 | 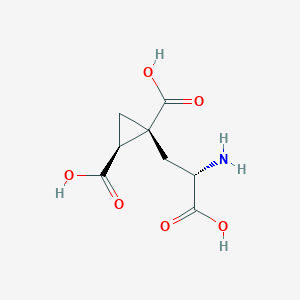 CHEMBL3787264 CHEMBL3787264 | C8H11NO6 | 217.177 | 7 / 4 | -4.0 | Yes |
| 522278 | 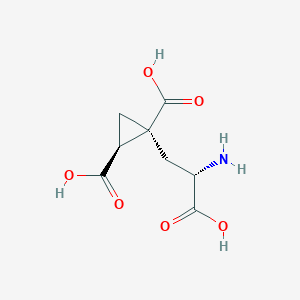 CHEMBL3786026 CHEMBL3786026 | C8H11NO6 | 217.177 | 7 / 4 | -4.0 | Yes |
| 522290 | 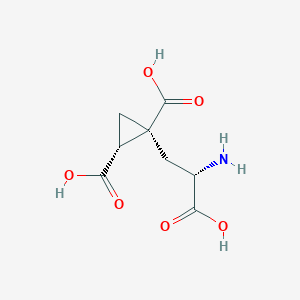 CHEMBL3786938 CHEMBL3786938 | C8H11NO6 | 217.177 | 7 / 4 | -4.0 | Yes |
| 27992 | 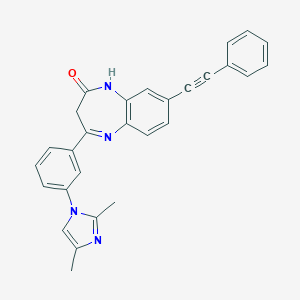 CHEMBL271319 CHEMBL271319 | C28H22N4O | 430.511 | 3 / 1 | 5.0 | Yes |
| 29326 | 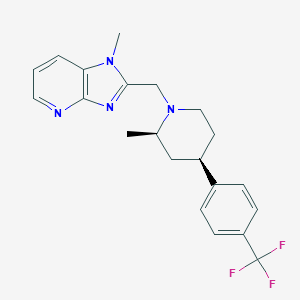 CHEMBL1774224 CHEMBL1774224 | C21H23F3N4 | 388.438 | 6 / 0 | 4.1 | Yes |
| 29403 | 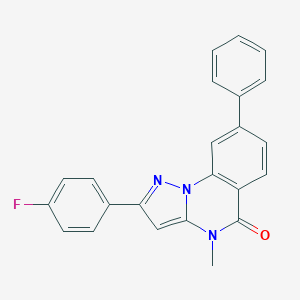 CHEMBL3286420 CHEMBL3286420 | C23H16FN3O | 369.399 | 3 / 0 | 4.7 | Yes |
| 30131 | 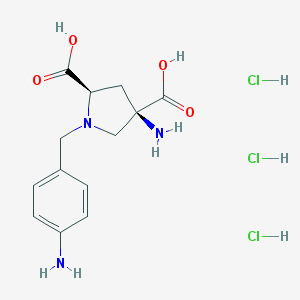 CHEMBL541050 CHEMBL541050 | C13H20Cl3N3O4 | 388.67 | 7 / 7 | N/A | No |
| 31462 | 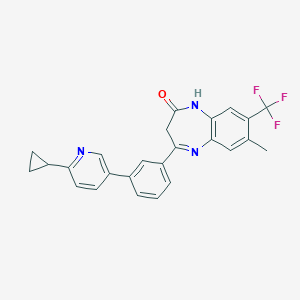 CHEMBL1629846 CHEMBL1629846 | C25H20F3N3O | 435.45 | 6 / 1 | 4.8 | Yes |
| 32184 | 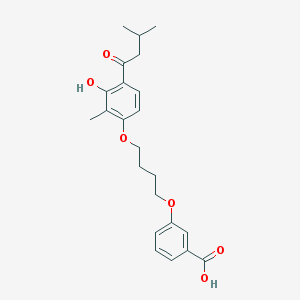 CHEMBL3287675 CHEMBL3287675 | C23H28O6 | 400.471 | 6 / 2 | 5.3 | No |
| 32245 | 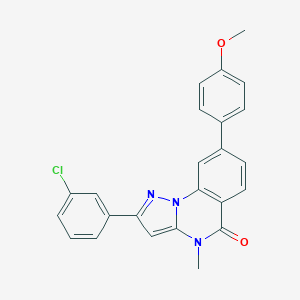 CHEMBL3288655 CHEMBL3288655 | C24H18ClN3O2 | 415.877 | 3 / 0 | 5.2 | No |
| 33519 | 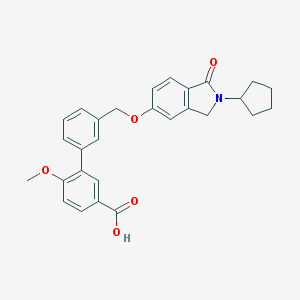 CHEMBL2179631 CHEMBL2179631 | C28H27NO5 | 457.526 | 5 / 1 | 4.8 | Yes |
| 35711 | 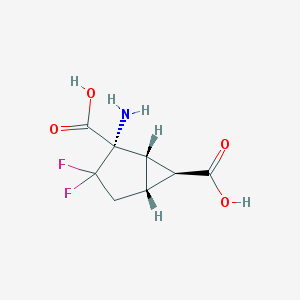 CHEMBL144201 CHEMBL144201 | C8H9F2NO4 | 221.16 | 7 / 3 | -2.9 | Yes |
| 36078 | 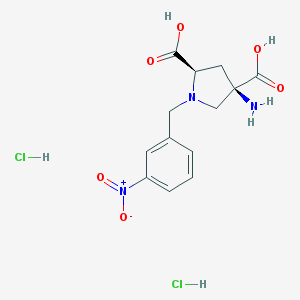 CHEMBL539760 CHEMBL539760 | C13H17Cl2N3O6 | 382.194 | 8 / 5 | N/A | N/A |
| 37242 | 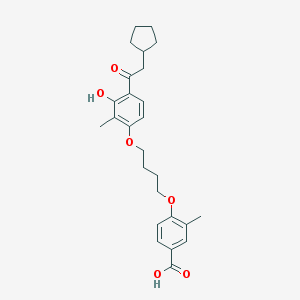 CHEMBL3287692 CHEMBL3287692 | C26H32O6 | 440.536 | 6 / 2 | 6.5 | No |
| 37459 | 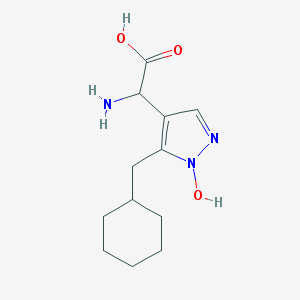 CHEMBL242344 CHEMBL242344 | C12H19N3O3 | 253.302 | 5 / 3 | -0.3 | Yes |
| 38397 | 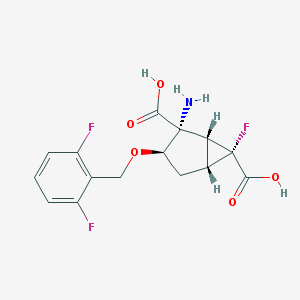 CHEMBL361051 CHEMBL361051 | C15H14F3NO5 | 345.274 | 9 / 3 | -1.7 | Yes |
| 459574 | 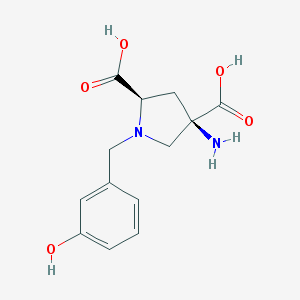 CHEMBL538487 CHEMBL538487 | C13H16N2O5 | 280.28 | 7 / 4 | -4.9 | Yes |
| 40235 | 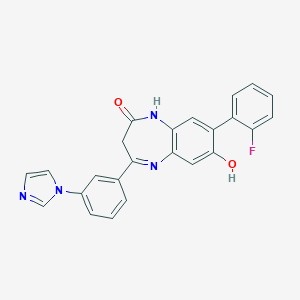 CHEMBL405895 CHEMBL405895 | C24H17FN4O2 | 412.424 | 5 / 2 | 3.5 | Yes |
| 42026 | 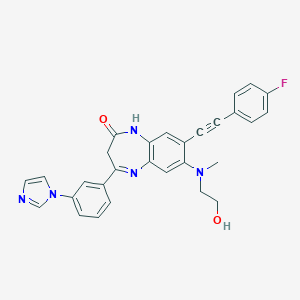 CHEMBL256424 CHEMBL256424 | C29H24FN5O2 | 493.542 | 6 / 2 | 3.7 | Yes |
| 42274 | 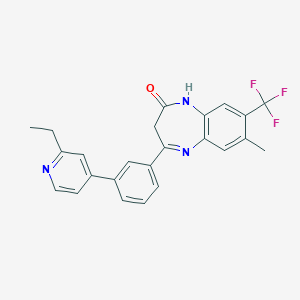 CHEMBL1631860 CHEMBL1631860 | C24H20F3N3O | 423.439 | 6 / 1 | 5.1 | No |
| 42652 | 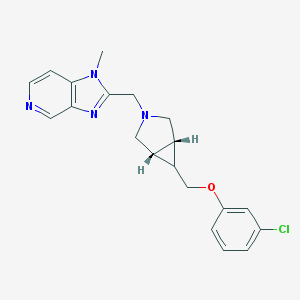 CHEMBL484578 CHEMBL484578 | C20H21ClN4O | 368.865 | 4 / 0 | 2.9 | Yes |
| 42853 | 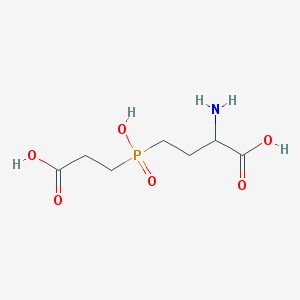 AC1MWWXC AC1MWWXC | C7H14NO6P | 239.164 | 7 / 4 | -4.8 | Yes |
| 43739 | 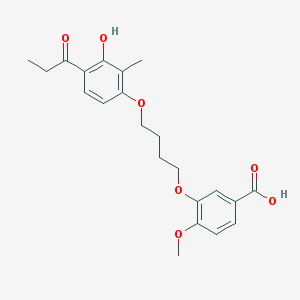 CHEMBL3287709 CHEMBL3287709 | C22H26O7 | 402.443 | 7 / 2 | 4.5 | Yes |
| 44373 | 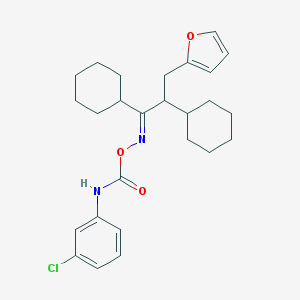 CHEMBL1209401 CHEMBL1209401 | C26H33ClN2O3 | 457.011 | 4 / 1 | 8.2 | No |
| 45465 | 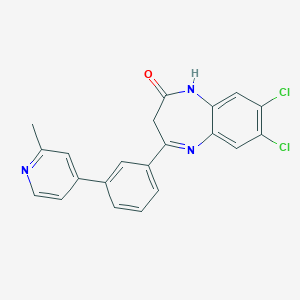 CHEMBL1629864 CHEMBL1629864 | C21H15Cl2N3O | 396.271 | 3 / 1 | 4.6 | Yes |
| 47139 | 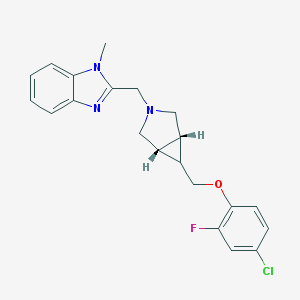 CHEMBL477375 CHEMBL477375 | C21H21ClFN3O | 385.867 | 4 / 0 | 4.0 | Yes |
| 48403 | 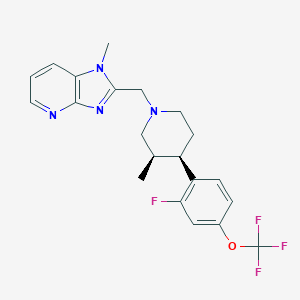 CHEMBL1774235 CHEMBL1774235 | C21H22F4N4O | 422.428 | 8 / 0 | 4.5 | Yes |
| 48405 | 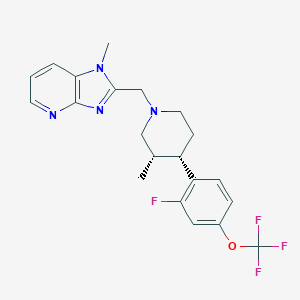 CHEMBL1774236 CHEMBL1774236 | C21H22F4N4O | 422.428 | 8 / 0 | 4.5 | Yes |
| 50546 | 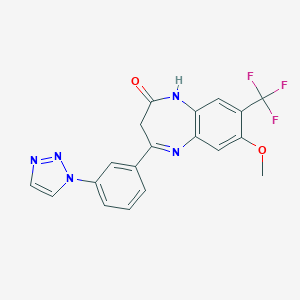 CHEMBL259340 CHEMBL259340 | C19H14F3N5O2 | 401.349 | 8 / 1 | 2.7 | Yes |
| 50635 | 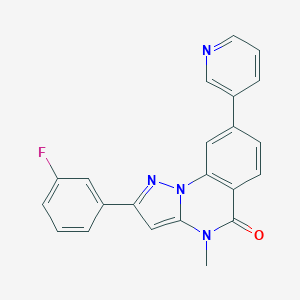 CHEMBL3288644 CHEMBL3288644 | C22H15FN4O | 370.387 | 4 / 0 | 3.6 | Yes |
| 52616 | 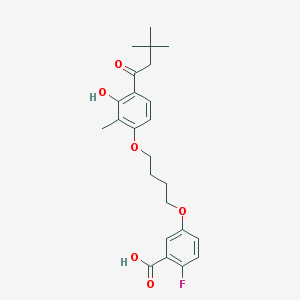 CHEMBL3287726 CHEMBL3287726 | C24H29FO6 | 432.488 | 7 / 2 | 5.8 | No |
| 53755 | 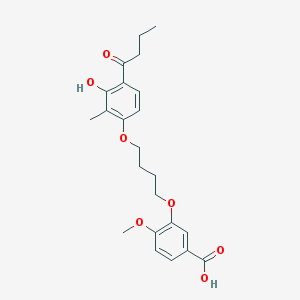 CHEMBL3287719 CHEMBL3287719 | C23H28O7 | 416.47 | 7 / 2 | 4.8 | Yes |
| 459733 | 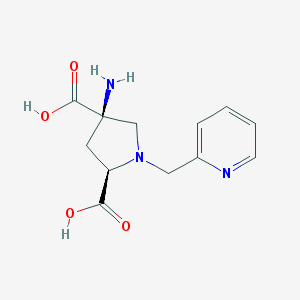 CHEMBL543811 CHEMBL543811 | C12H15N3O4 | 265.269 | 7 / 3 | -5.6 | Yes |
| 57002 | 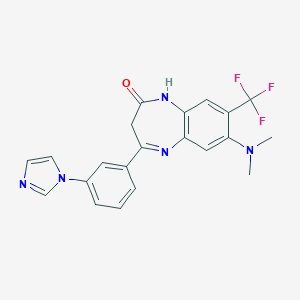 CHEMBL263649 CHEMBL263649 | C21H18F3N5O | 413.404 | 7 / 1 | 3.2 | Yes |
| 57284 | 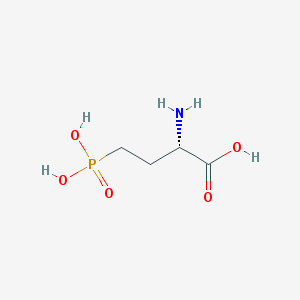 L-AP4 L-AP4 | C4H10NO5P | 183.1 | 6 / 4 | -5.5 | Yes |
| 58687 | 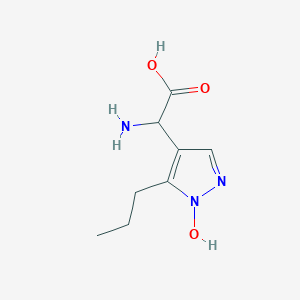 CHEMBL389585 CHEMBL389585 | C8H13N3O3 | 199.21 | 5 / 3 | -2.1 | Yes |
| 58964 | 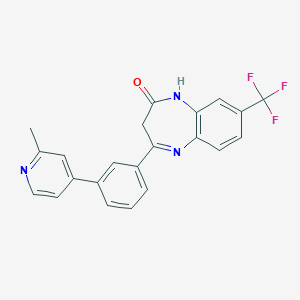 CHEMBL1629865 CHEMBL1629865 | C22H16F3N3O | 395.385 | 6 / 1 | 4.3 | Yes |
| 59249 | 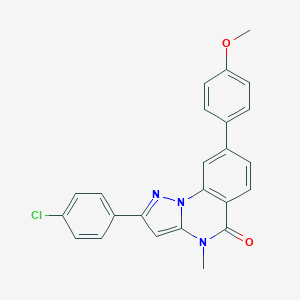 CHEMBL3288666 CHEMBL3288666 | C24H18ClN3O2 | 415.877 | 3 / 0 | 5.2 | No |
| 59330 | 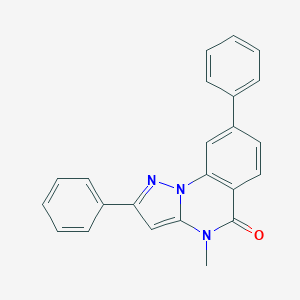 CHEMBL3288637 CHEMBL3288637 | C23H17N3O | 351.409 | 2 / 0 | 4.6 | Yes |
| 59677 | 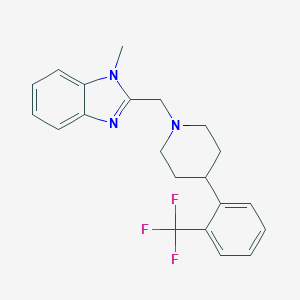 CHEMBL604467 CHEMBL604467 | C21H22F3N3 | 373.423 | 5 / 0 | 4.4 | Yes |
| 60112 | 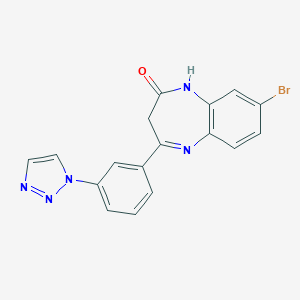 CHEMBL260343 CHEMBL260343 | C17H12BrN5O | 382.221 | 4 / 1 | 2.5 | Yes |
| 60277 | 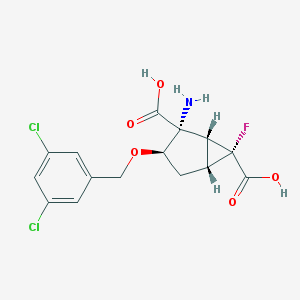 CHEMBL363980 CHEMBL363980 | C15H14Cl2FNO5 | 378.177 | 7 / 3 | -0.7 | Yes |
| 61154 | 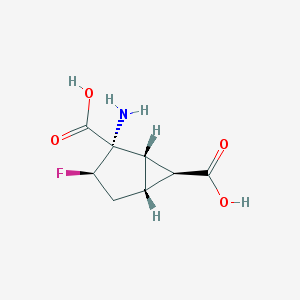 CHEMBL341888 CHEMBL341888 | C8H10FNO4 | 203.169 | 6 / 3 | -3.2 | Yes |
| 61155 | 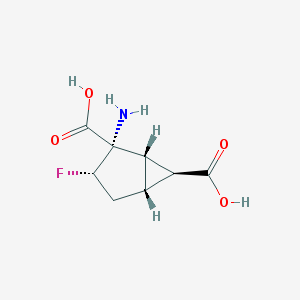 CHEMBL334014 CHEMBL334014 | C8H10FNO4 | 203.169 | 6 / 3 | -3.2 | Yes |
| 61165 | 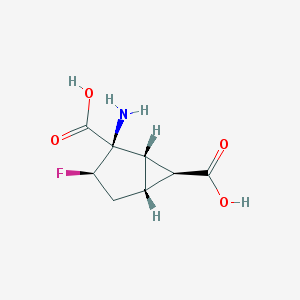 CHEMBL144678 CHEMBL144678 | C8H10FNO4 | 203.169 | 6 / 3 | -3.2 | Yes |
| 61456 | 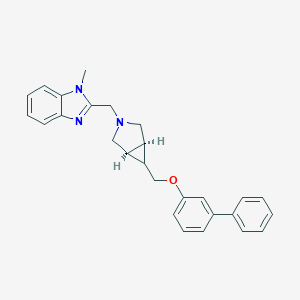 CHEMBL476791 CHEMBL476791 | C27H27N3O | 409.533 | 3 / 0 | 4.9 | Yes |
| 62351 | 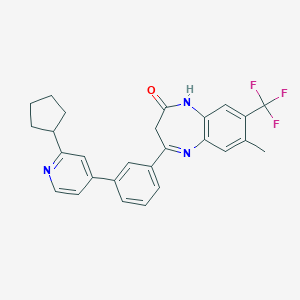 CHEMBL1631865 CHEMBL1631865 | C27H24F3N3O | 463.504 | 6 / 1 | 5.9 | No |
| 62812 | 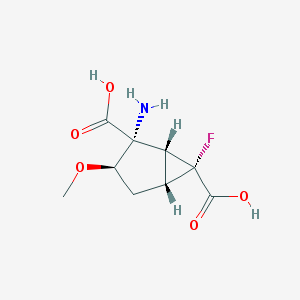 CHEMBL183956 CHEMBL183956 | C9H12FNO5 | 233.195 | 7 / 3 | -3.4 | Yes |
| 62814 | 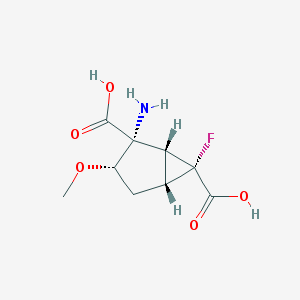 CHEMBL185135 CHEMBL185135 | C9H12FNO5 | 233.195 | 7 / 3 | -3.4 | Yes |
| 63091 | 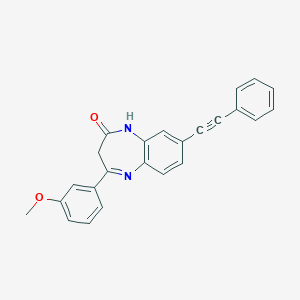 CHEMBL400018 CHEMBL400018 | C24H18N2O2 | 366.42 | 3 / 1 | 4.4 | Yes |
| 459792 | 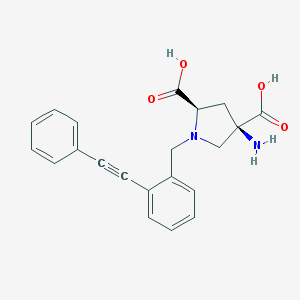 CHEMBL539516 CHEMBL539516 | C21H20N2O4 | 364.401 | 6 / 3 | -2.5 | Yes |
| 63606 | 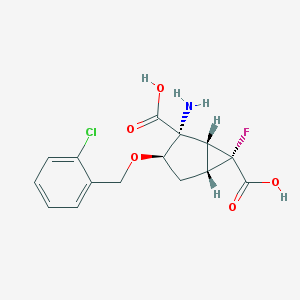 CHEMBL362092 CHEMBL362092 | C15H15ClFNO5 | 343.735 | 7 / 3 | -1.3 | Yes |
| 64091 | 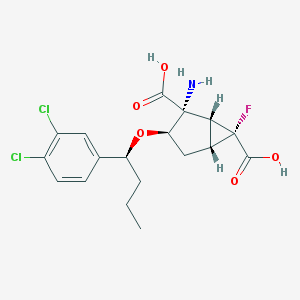 CHEMBL181710 CHEMBL181710 | C18H20Cl2FNO5 | 420.258 | 7 / 3 | 0.6 | Yes |
| 64092 | 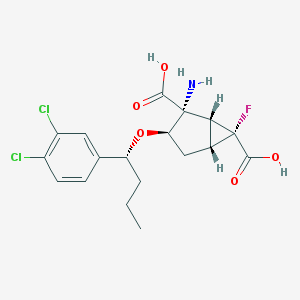 CHEMBL365680 CHEMBL365680 | C18H20Cl2FNO5 | 420.258 | 7 / 3 | 0.6 | Yes |
| 553564 | 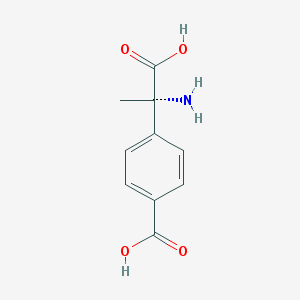 (S)-MCPG (S)-MCPG | C10H11NO4 | 209.201 | 5 / 3 | -2.0 | Yes |
| 64228 | 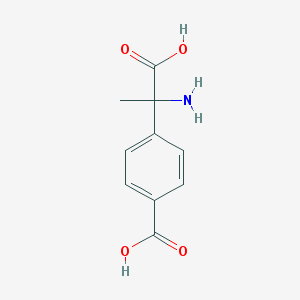 (RS)-MCPG (RS)-MCPG | C10H11NO4 | 209.201 | 5 / 3 | -2.0 | Yes |
| 65204 | 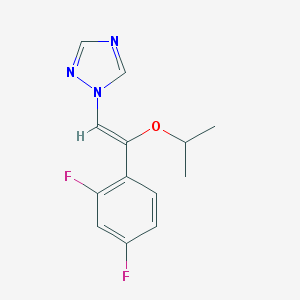 CHEMBL69869 CHEMBL69869 | C13H13F2N3O | 265.264 | 5 / 0 | 2.9 | Yes |
| 65347 | 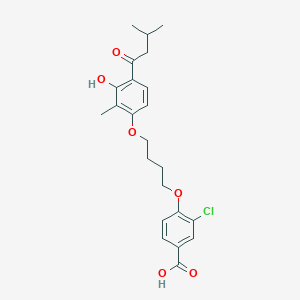 CHEMBL3287697 CHEMBL3287697 | C23H27ClO6 | 434.913 | 6 / 2 | 5.9 | No |
| 66431 | 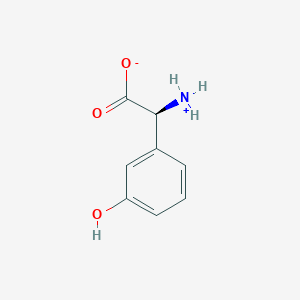 CID 40428795 CID 40428795 | C8H9NO3 | 167.164 | 3 / 2 | -1.4 | Yes |
| 459826 | 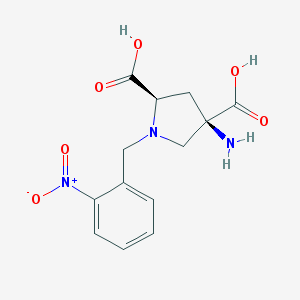 CHEMBL544508 CHEMBL544508 | C13H15N3O6 | 309.278 | 8 / 3 | -4.7 | Yes |
| 66649 | 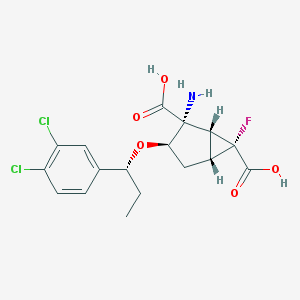 CHEMBL184983 CHEMBL184983 | C17H18Cl2FNO5 | 406.231 | 7 / 3 | 0.3 | Yes |
| 66650 | 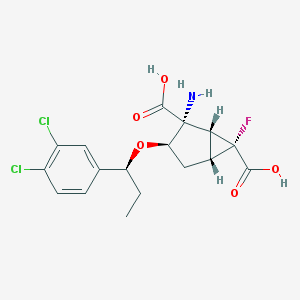 CHEMBL185423 CHEMBL185423 | C17H18Cl2FNO5 | 406.231 | 7 / 3 | 0.3 | Yes |
| 68413 | 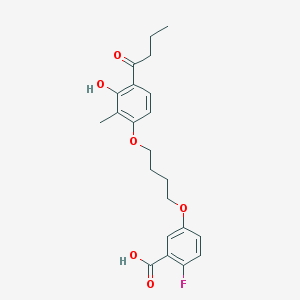 CHEMBL3287721 CHEMBL3287721 | C22H25FO6 | 404.434 | 7 / 2 | 5.0 | Yes |
| 68440 | 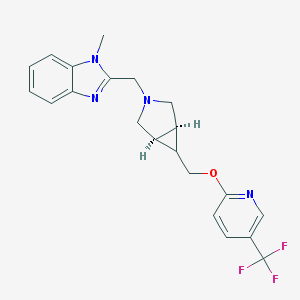 CHEMBL515034 CHEMBL515034 | C21H21F3N4O | 402.421 | 7 / 0 | 3.4 | Yes |
| 69278 | 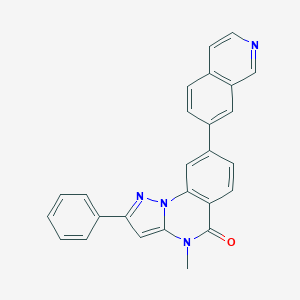 CHEMBL3288680 CHEMBL3288680 | C26H18N4O | 402.457 | 3 / 0 | 4.8 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417