

You can:
| Name | Taste receptor type 2 member 8 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | TAS2R8 |
| Synonym | T2R8 TAS2R8 Taste receptor family B member 5 taste receptor, type 2, member 8 TRB5 |
| Disease | N/A |
| Length | 309 |
| Amino acid sequence | MFSPADNIFIILITGEFILGILGNGYIALVNWIDWIKKKKISTVDYILTNLVIARICLISVMVVNGIVIVLNPDVYTKNKQQIVIFTFWTFANYLNMWITTCLNVFYFLKIASSSHPLFLWLKWKIDMVVHWILLGCFAISLLVSLIAAIVLSCDYRFHAIAKHKRNITEMFHVSKIPYFEPLTLFNLFAIVPFIVSLISFFLLVRSLWRHTKQIKLYATGSRDPSTEVHVRAIKTMTSFIFFFFLYYISSILMTFSYLMTKYKLAVEFGEIAAILYPLGHSLILIVLNNKLRQTFVRMLTCRKIACMI |
| UniProt | Q9NYW2 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q9NYW2 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q9NYW2. |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL3988599 |
| IUPHAR | N/A |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 519717 | 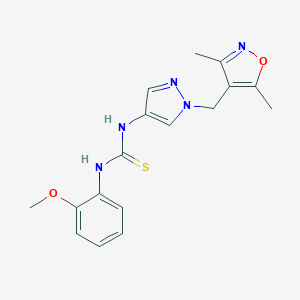 SCHEMBL1278959 SCHEMBL1278959 | C17H19N5O2S | 357.432 | 5 / 2 | 2.2 | Yes |
| 519719 | 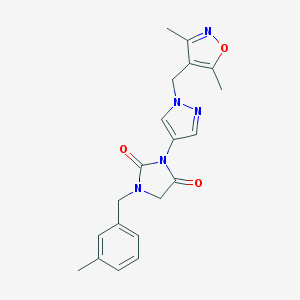 SCHEMBL1278797 SCHEMBL1278797 | C20H21N5O3 | 379.42 | 5 / 0 | 1.9 | Yes |
| 519720 | 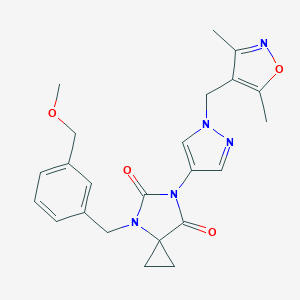 SCHEMBL1279727 SCHEMBL1279727 | C23H25N5O4 | 435.484 | 6 / 0 | 1.5 | Yes |
| 519725 | 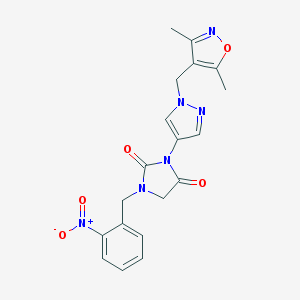 SCHEMBL1278923 SCHEMBL1278923 | C19H18N6O5 | 410.39 | 7 / 0 | 1.4 | Yes |
| 536113 | 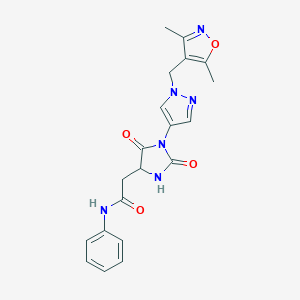 SCHEMBL1279180 SCHEMBL1279180 | C20H20N6O4 | 408.418 | 6 / 2 | 0.7 | Yes |
| 519730 | 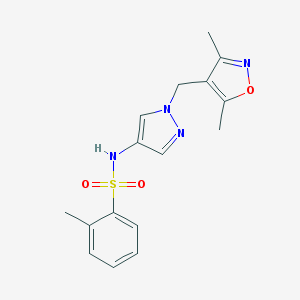 SCHEMBL1614927 SCHEMBL1614927 | C16H18N4O3S | 346.405 | 6 / 1 | 2.0 | Yes |
| 536295 | 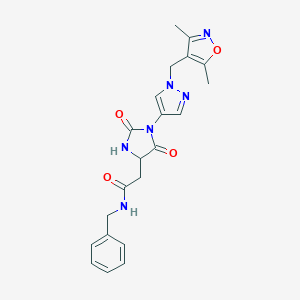 SCHEMBL1279594 SCHEMBL1279594 | C21H22N6O4 | 422.445 | 6 / 2 | 0.6 | Yes |
| 519755 | 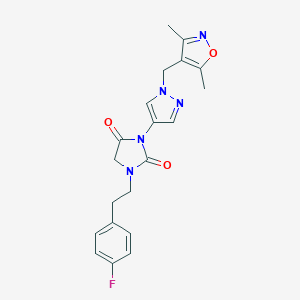 SCHEMBL1279016 SCHEMBL1279016 | C20H20FN5O3 | 397.41 | 6 / 0 | 2.1 | Yes |
| 519761 | 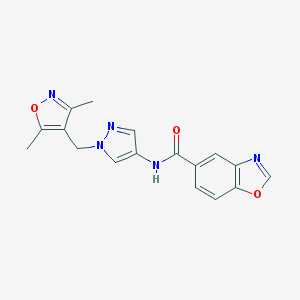 SCHEMBL1278560 SCHEMBL1278560 | C17H15N5O3 | 337.339 | 6 / 1 | 1.8 | Yes |
| 519767 | 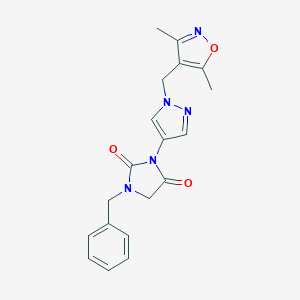 SCHEMBL1278762 SCHEMBL1278762 | C19H19N5O3 | 365.393 | 5 / 0 | 1.5 | Yes |
| 519772 | 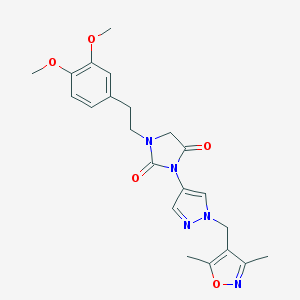 SCHEMBL1279943 SCHEMBL1279943 | C22H25N5O5 | 439.472 | 7 / 0 | 1.9 | Yes |
| 519784 | 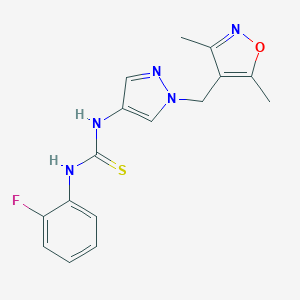 SCHEMBL1279101 SCHEMBL1279101 | C16H16FN5OS | 345.396 | 5 / 2 | 2.3 | Yes |
| 519785 | 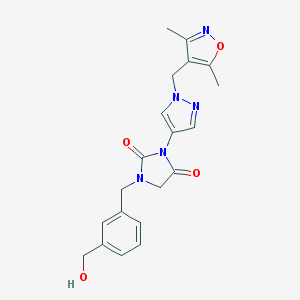 SCHEMBL1279313 SCHEMBL1279313 | C20H21N5O4 | 395.419 | 6 / 1 | 0.6 | Yes |
| 519792 | 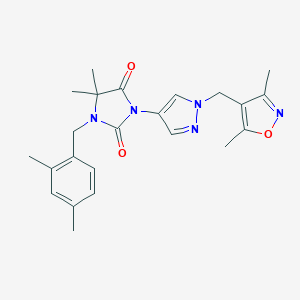 SCHEMBL1616623 SCHEMBL1616623 | C23H27N5O3 | 421.501 | 5 / 0 | 2.8 | Yes |
| 519797 | 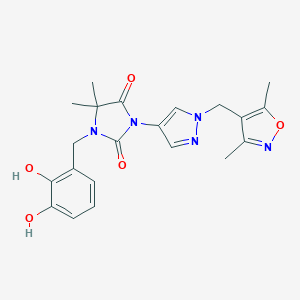 SCHEMBL1615598 SCHEMBL1615598 | C21H23N5O5 | 425.445 | 7 / 2 | 1.4 | Yes |
| 519798 | 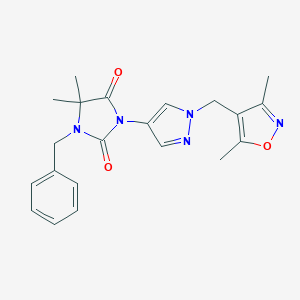 SCHEMBL1615909 SCHEMBL1615909 | C21H23N5O3 | 393.447 | 5 / 0 | 2.1 | Yes |
| 519806 | 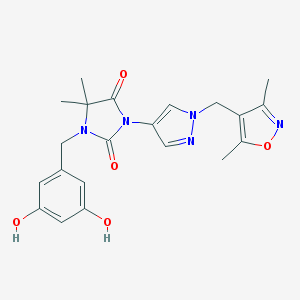 SCHEMBL1616554 SCHEMBL1616554 | C21H23N5O5 | 425.445 | 7 / 2 | 1.4 | Yes |
| 519813 | 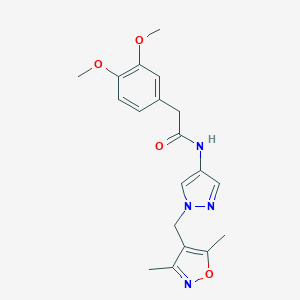 SCHEMBL1615834 SCHEMBL1615834 | C19H22N4O4 | 370.409 | 6 / 1 | 1.8 | Yes |
| 519825 | 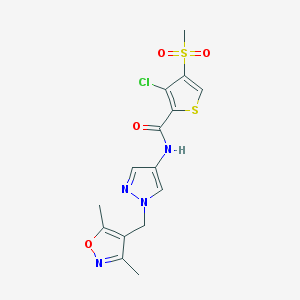 AC1N8BT6 AC1N8BT6 | C15H15ClN4O4S2 | 414.879 | 7 / 1 | 1.8 | Yes |
| 519830 | 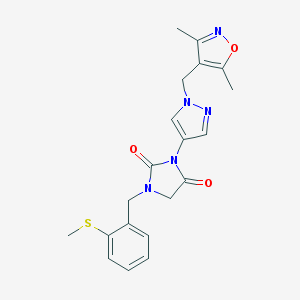 SCHEMBL1278994 SCHEMBL1278994 | C20H21N5O3S | 411.48 | 6 / 0 | 2.0 | Yes |
| 519831 | 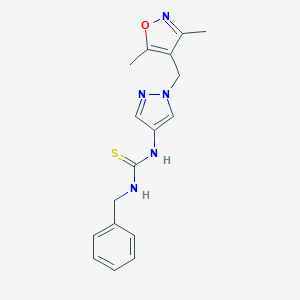 SCHEMBL1278840 SCHEMBL1278840 | C17H19N5OS | 341.433 | 4 / 2 | 2.2 | Yes |
| 519832 | 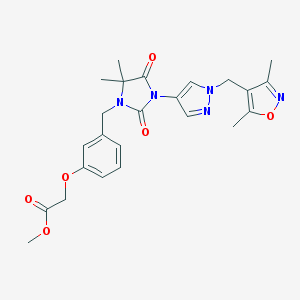 SCHEMBL1616720 SCHEMBL1616720 | C24H27N5O6 | 481.509 | 8 / 0 | 2.0 | Yes |
| 519834 | 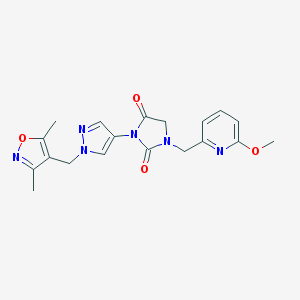 SCHEMBL1279356 SCHEMBL1279356 | C19H20N6O4 | 396.407 | 7 / 0 | 0.8 | Yes |
| 519842 | 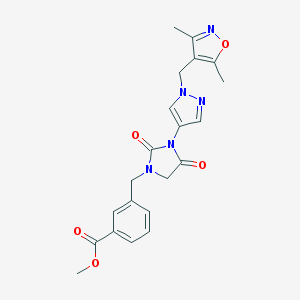 SCHEMBL1279616 SCHEMBL1279616 | C21H21N5O5 | 423.429 | 7 / 0 | 1.4 | Yes |
| 519847 | 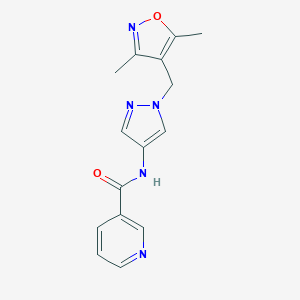 SCHEMBL1278991 SCHEMBL1278991 | C15H15N5O2 | 297.318 | 5 / 1 | 0.8 | Yes |
| 519853 | 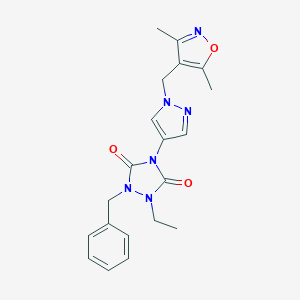 SCHEMBL1279646 SCHEMBL1279646 | C20H22N6O3 | 394.435 | 5 / 0 | 2.1 | Yes |
| 536866 | 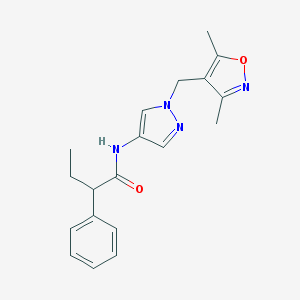 SCHEMBL1615950 SCHEMBL1615950 | C19H22N4O2 | 338.411 | 4 / 1 | 2.8 | Yes |
| 519861 | 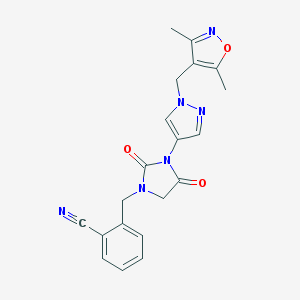 SCHEMBL1279176 SCHEMBL1279176 | C20H18N6O3 | 390.403 | 6 / 0 | 1.2 | Yes |
| 519863 | 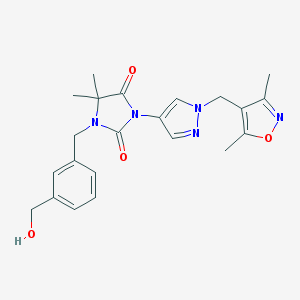 SCHEMBL1279520 SCHEMBL1279520 | C22H25N5O4 | 423.473 | 6 / 1 | 1.2 | Yes |
| 536999 | 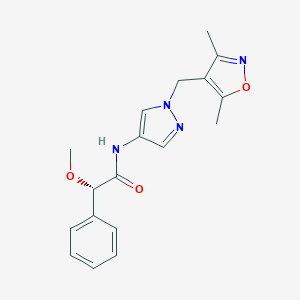 SCHEMBL1279389 SCHEMBL1279389 | C18H20N4O3 | 340.383 | 5 / 1 | 1.8 | Yes |
| 558469 | 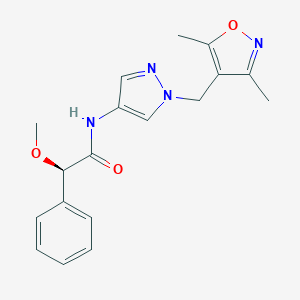 SCHEMBL1279211 SCHEMBL1279211 | C18H20N4O3 | 340.383 | 5 / 1 | 1.8 | Yes |
| 537000 | 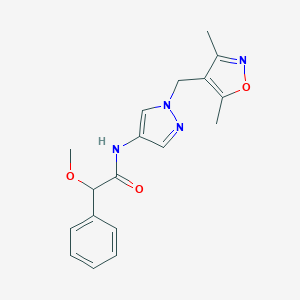 SCHEMBL1278930 SCHEMBL1278930 | C18H20N4O3 | 340.383 | 5 / 1 | 1.8 | Yes |
| 519866 | 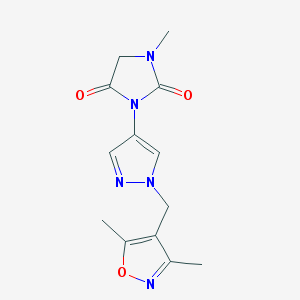 SCHEMBL1278701 SCHEMBL1278701 | C13H15N5O3 | 289.295 | 5 / 0 | 0.0 | Yes |
| 519869 | 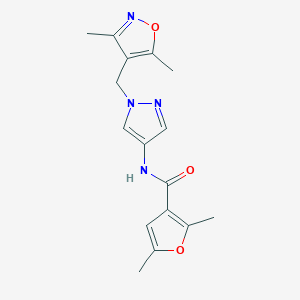 SCHEMBL1279406 SCHEMBL1279406 | C16H18N4O3 | 314.345 | 5 / 1 | 1.8 | Yes |
| 519873 | 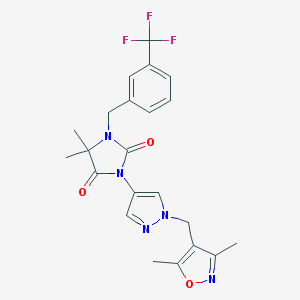 SCHEMBL1616615 SCHEMBL1616615 | C22H22F3N5O3 | 461.445 | 8 / 0 | 3.0 | Yes |
| 519884 | 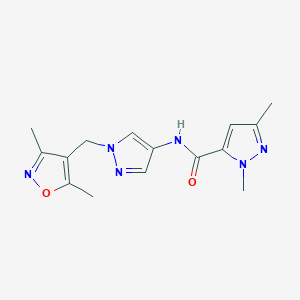 SCHEMBL1279201 SCHEMBL1279201 | C15H18N6O2 | 314.349 | 5 / 1 | 1.0 | Yes |
| 519901 | 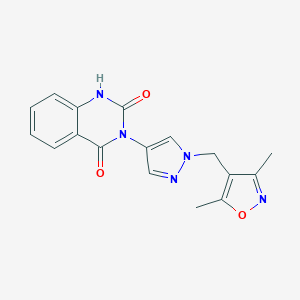 SCHEMBL1279621 SCHEMBL1279621 | C17H15N5O3 | 337.339 | 5 / 1 | 1.3 | Yes |
| 519906 | 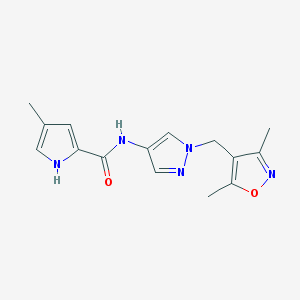 SCHEMBL1279073 SCHEMBL1279073 | C15H17N5O2 | 299.334 | 4 / 2 | 1.4 | Yes |
| 519908 | 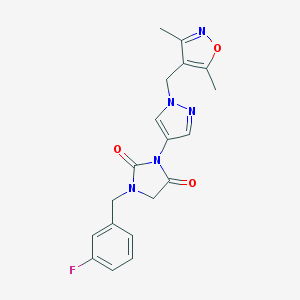 SCHEMBL1279276 SCHEMBL1279276 | C19H18FN5O3 | 383.383 | 6 / 0 | 1.6 | Yes |
| 519912 | 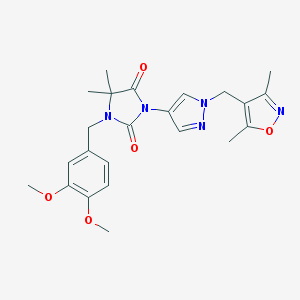 SCHEMBL1279508 SCHEMBL1279508 | C23H27N5O5 | 453.499 | 7 / 0 | 2.1 | Yes |
| 519925 | 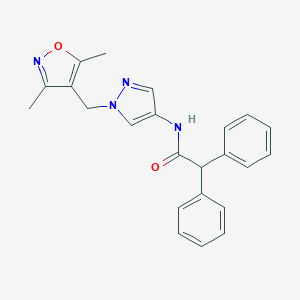 SCHEMBL1615713 SCHEMBL1615713 | C23H22N4O2 | 386.455 | 4 / 1 | 3.5 | Yes |
| 519935 | 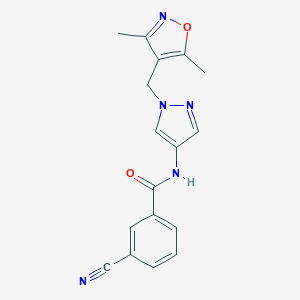 SCHEMBL1278611 SCHEMBL1278611 | C17H15N5O2 | 321.34 | 5 / 1 | 1.6 | Yes |
| 519943 | 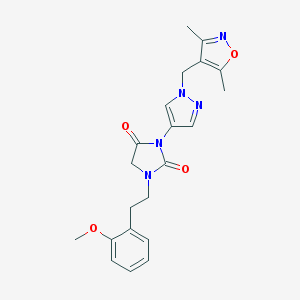 SCHEMBL1279766 SCHEMBL1279766 | C21H23N5O4 | 409.446 | 6 / 0 | 2.0 | Yes |
| 519944 | 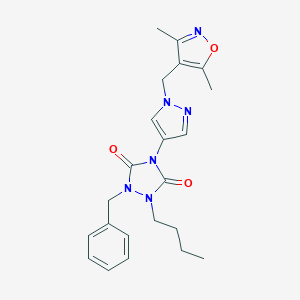 SCHEMBL1279293 SCHEMBL1279293 | C22H26N6O3 | 422.489 | 5 / 0 | 3.0 | Yes |
| 519945 |  SCHEMBL1278938 SCHEMBL1278938 | C22H26N6O3 | 422.489 | 6 / 0 | 1.4 | Yes |
| 519962 |  SCHEMBL3670969 SCHEMBL3670969 | C23H27N5O5 | 453.499 | 7 / 0 | 2.1 | Yes |
| 519967 |  SCHEMBL1615607 SCHEMBL1615607 | C22H25N5O3 | 407.474 | 5 / 0 | 2.5 | Yes |
| 519968 |  SCHEMBL1278714 SCHEMBL1278714 | C19H18FN5O3 | 383.383 | 6 / 0 | 1.6 | Yes |
| 519973 | 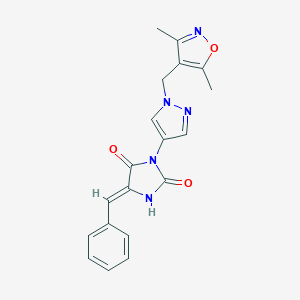 SCHEMBL1278924 SCHEMBL1278924 | C19H17N5O3 | 363.377 | 5 / 1 | 2.0 | Yes |
| 519975 | 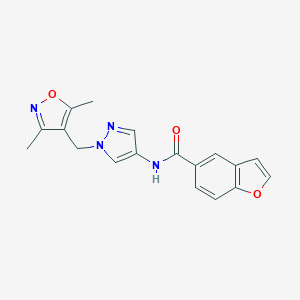 SCHEMBL1279235 SCHEMBL1279235 | C18H16N4O3 | 336.351 | 5 / 1 | 2.3 | Yes |
| 519984 | 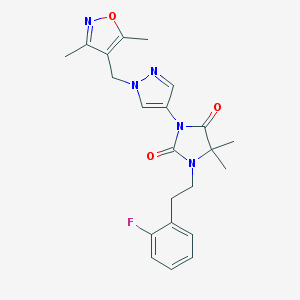 SCHEMBL1616590 SCHEMBL1616590 | C22H24FN5O3 | 425.464 | 6 / 0 | 2.7 | Yes |
| 519986 | 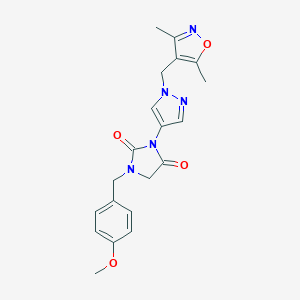 SCHEMBL1279167 SCHEMBL1279167 | C20H21N5O4 | 395.419 | 6 / 0 | 1.5 | Yes |
| 519990 | 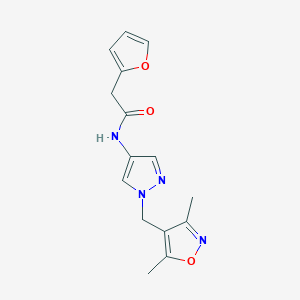 SCHEMBL1615968 SCHEMBL1615968 | C15H16N4O3 | 300.318 | 5 / 1 | 0.9 | Yes |
| 519997 | 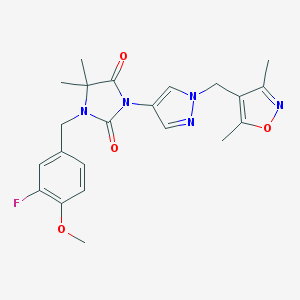 SCHEMBL1616185 SCHEMBL1616185 | C22H24FN5O4 | 441.463 | 7 / 0 | 2.2 | Yes |
| 559438 | 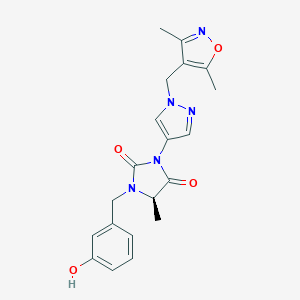 SCHEMBL1279395 SCHEMBL1279395 | C20H21N5O4 | 395.419 | 6 / 1 | 1.6 | Yes |
| 537650 | 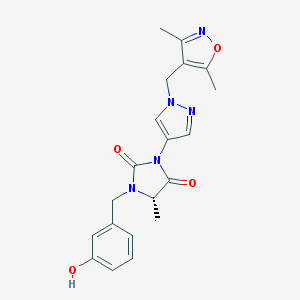 SCHEMBL1279181 SCHEMBL1279181 | C20H21N5O4 | 395.419 | 6 / 1 | 1.6 | Yes |
| 537654 | 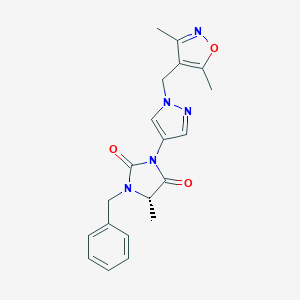 SCHEMBL1279205 SCHEMBL1279205 | C20H21N5O3 | 379.42 | 5 / 0 | 1.9 | Yes |
| 559452 | 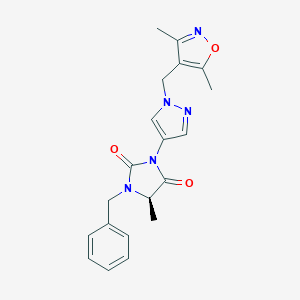 SCHEMBL1279690 SCHEMBL1279690 | C20H21N5O3 | 379.42 | 5 / 0 | 1.9 | Yes |
| 537655 | 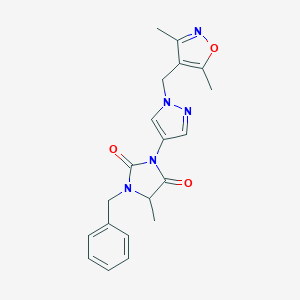 SCHEMBL1279209 SCHEMBL1279209 | C20H21N5O3 | 379.42 | 5 / 0 | 1.9 | Yes |
| 520009 | 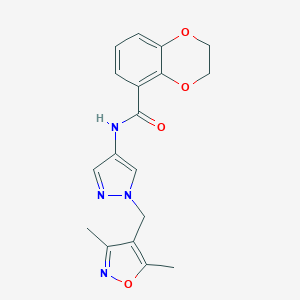 SCHEMBL1652683 SCHEMBL1652683 | C18H18N4O4 | 354.366 | 6 / 1 | 1.6 | Yes |
| 520011 | 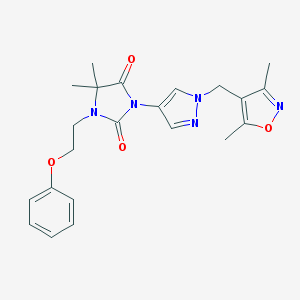 SCHEMBL1279617 SCHEMBL1279617 | C22H25N5O4 | 423.473 | 6 / 0 | 2.3 | Yes |
| 520012 | 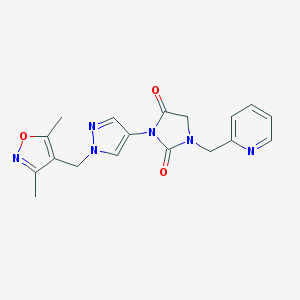 SCHEMBL1279343 SCHEMBL1279343 | C18H18N6O3 | 366.381 | 6 / 0 | 0.5 | Yes |
| 520017 | 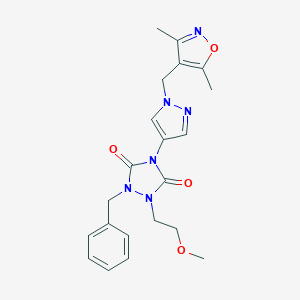 SCHEMBL1279790 SCHEMBL1279790 | C21H24N6O4 | 424.461 | 6 / 0 | 1.6 | Yes |
| 537780 | 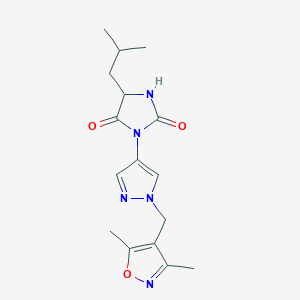 SCHEMBL1278689 SCHEMBL1278689 | C16H21N5O3 | 331.376 | 5 / 1 | 1.6 | Yes |
| 520018 | 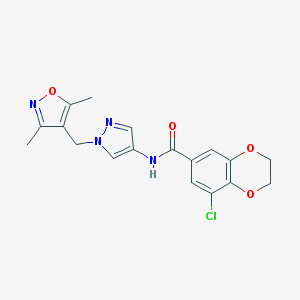 SCHEMBL1649359 SCHEMBL1649359 | C18H17ClN4O4 | 388.808 | 6 / 1 | 2.3 | Yes |
| 520026 | 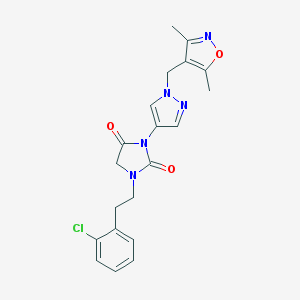 SCHEMBL1279600 SCHEMBL1279600 | C20H20ClN5O3 | 413.862 | 5 / 0 | 2.6 | Yes |
| 520027 | 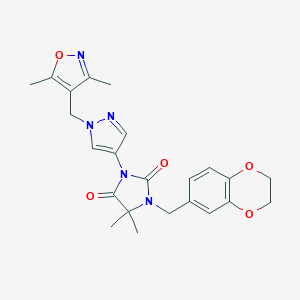 SCHEMBL1615620 SCHEMBL1615620 | C23H25N5O5 | 451.483 | 7 / 0 | 1.8 | Yes |
| 520030 | 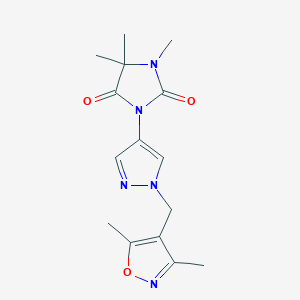 SCHEMBL1279815 SCHEMBL1279815 | C15H19N5O3 | 317.349 | 5 / 0 | 0.6 | Yes |
| 537898 |  SCHEMBL1279076 SCHEMBL1279076 | C18H23N5O3 | 357.414 | 5 / 1 | 2.4 | Yes |
| 520064 |  SCHEMBL1279415 SCHEMBL1279415 | C19H25N5O3 | 371.441 | 5 / 0 | 2.4 | Yes |
| 520070 |  SCHEMBL1279261 SCHEMBL1279261 | C14H15N5O2 | 285.307 | 4 / 2 | 1.0 | Yes |
| 520081 |  SCHEMBL1279855 SCHEMBL1279855 | C20H25N7O3 | 411.466 | 6 / 0 | 0.9 | Yes |
| 520095 | 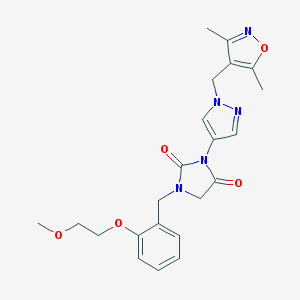 SCHEMBL1279611 SCHEMBL1279611 | C22H25N5O5 | 439.472 | 7 / 0 | 1.3 | Yes |
| 520102 | 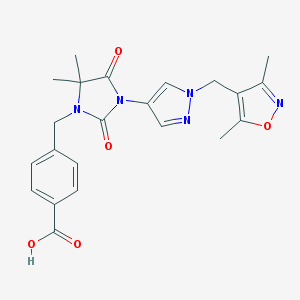 SCHEMBL1616647 SCHEMBL1616647 | C22H23N5O5 | 437.456 | 7 / 1 | 1.6 | Yes |
| 538424 | 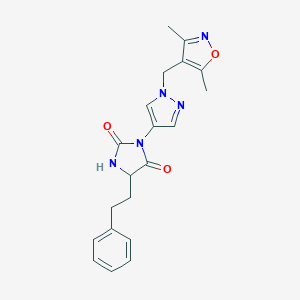 SCHEMBL1279090 SCHEMBL1279090 | C20H21N5O3 | 379.42 | 5 / 1 | 2.2 | Yes |
| 538427 | 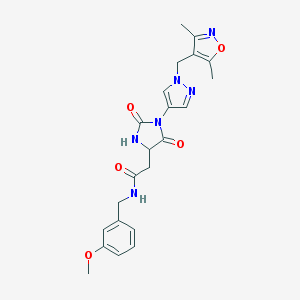 SCHEMBL1279138 SCHEMBL1279138 | C22H24N6O5 | 452.471 | 7 / 2 | 0.6 | Yes |
| 560387 | 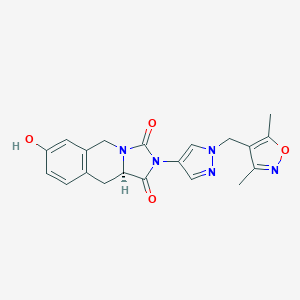 SCHEMBL1279983 SCHEMBL1279983 | C20H19N5O4 | 393.403 | 6 / 1 | 1.2 | Yes |
| 538428 | 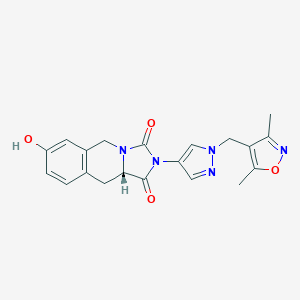 SCHEMBL1280225 SCHEMBL1280225 | C20H19N5O4 | 393.403 | 6 / 1 | 1.2 | Yes |
| 538519 | 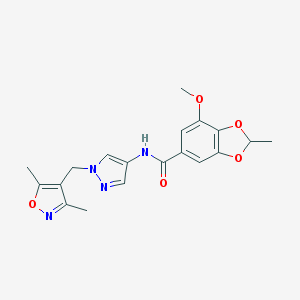 SCHEMBL1278850 SCHEMBL1278850 | C19H20N4O5 | 384.392 | 7 / 1 | 2.1 | Yes |
| 520141 | 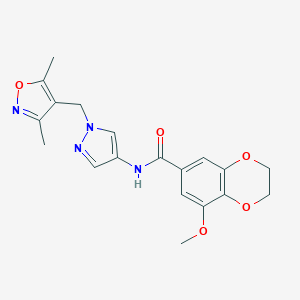 SCHEMBL1279043 SCHEMBL1279043 | C19H20N4O5 | 384.392 | 7 / 1 | 1.6 | Yes |
| 520143 | 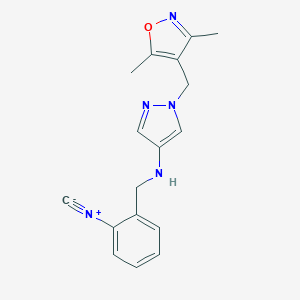 SCHEMBL1616196 SCHEMBL1616196 | C17H17N5O | 307.357 | 5 / 1 | 2.3 | Yes |
| 520162 | 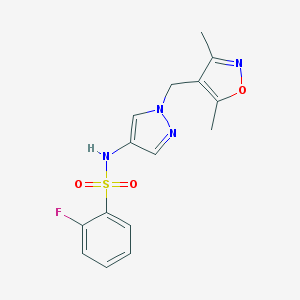 SCHEMBL1615384 SCHEMBL1615384 | C15H15FN4O3S | 350.368 | 7 / 1 | 1.7 | Yes |
| 520176 | 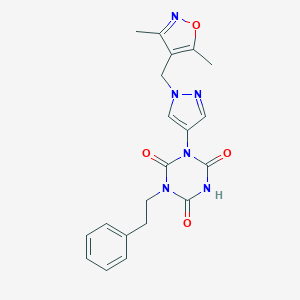 SCHEMBL1279302 SCHEMBL1279302 | C20H20N6O4 | 408.418 | 6 / 1 | 1.9 | Yes |
| 520179 | 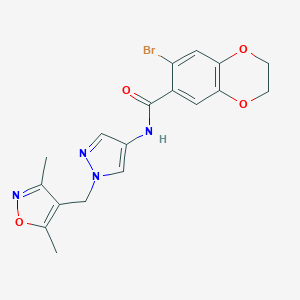 SCHEMBL1650803 SCHEMBL1650803 | C18H17BrN4O4 | 433.262 | 6 / 1 | 2.3 | Yes |
| 520201 | 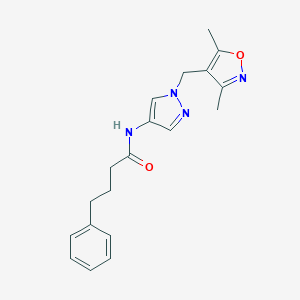 SCHEMBL1614872 SCHEMBL1614872 | C19H22N4O2 | 338.411 | 4 / 1 | 2.5 | Yes |
| 520205 | 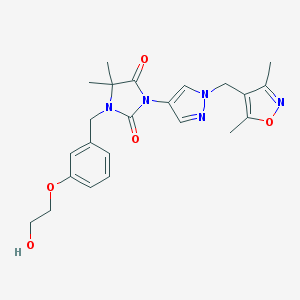 SCHEMBL1615626 SCHEMBL1615626 | C23H27N5O5 | 453.499 | 7 / 1 | 1.4 | Yes |
| 538990 | 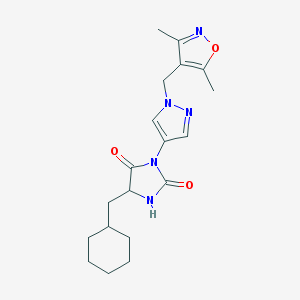 SCHEMBL1279237 SCHEMBL1279237 | C19H25N5O3 | 371.441 | 5 / 1 | 3.0 | Yes |
| 520215 | 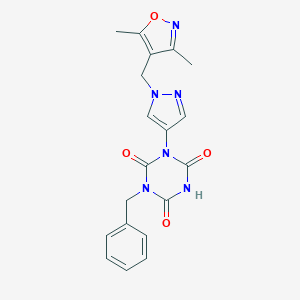 SCHEMBL1279105 SCHEMBL1279105 | C19H18N6O4 | 394.391 | 6 / 1 | 1.4 | Yes |
| 520217 | 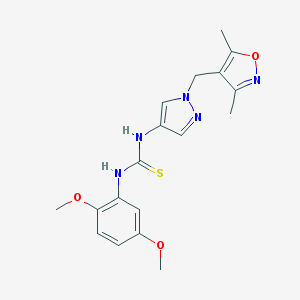 AC1MVSHT AC1MVSHT | C18H21N5O3S | 387.458 | 6 / 2 | 2.2 | Yes |
| 520221 | 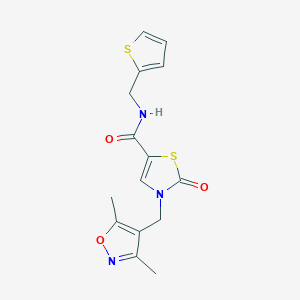 SCHEMBL1278962 SCHEMBL1278962 | C15H15N3O3S2 | 349.423 | 6 / 1 | 1.9 | Yes |
| 539089 | 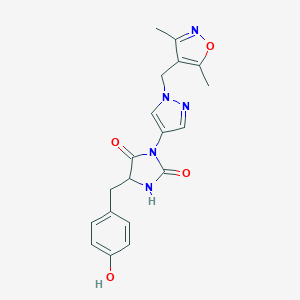 SCHEMBL1279666 SCHEMBL1279666 | C19H19N5O4 | 381.392 | 6 / 2 | 1.5 | Yes |
| 520227 | 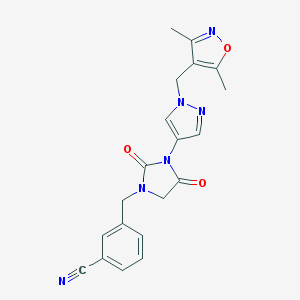 SCHEMBL1279134 SCHEMBL1279134 | C20H18N6O3 | 390.403 | 6 / 0 | 1.2 | Yes |
| 520234 | 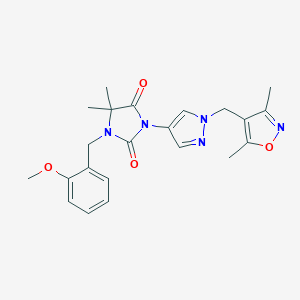 SCHEMBL1279359 SCHEMBL1279359 | C22H25N5O4 | 423.473 | 6 / 0 | 2.1 | Yes |
| 520239 | 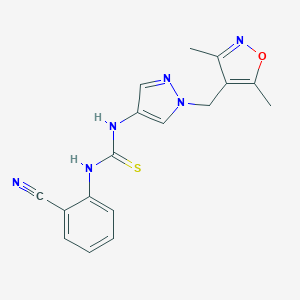 SCHEMBL1279597 SCHEMBL1279597 | C17H16N6OS | 352.416 | 5 / 2 | 2.5 | Yes |
| 520242 | 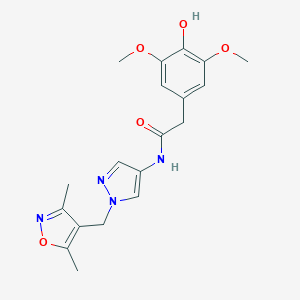 SCHEMBL1278315 SCHEMBL1278315 | C19H22N4O5 | 386.408 | 7 / 2 | 1.4 | Yes |
| 520259 | 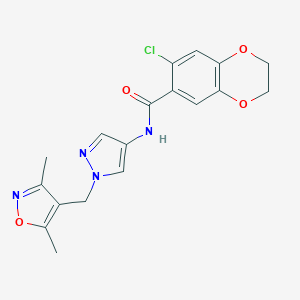 SCHEMBL1650405 SCHEMBL1650405 | C18H17ClN4O4 | 388.808 | 6 / 1 | 2.3 | Yes |
| 520261 | 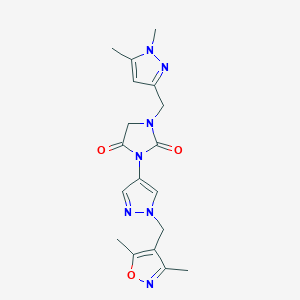 SCHEMBL1279273 SCHEMBL1279273 | C18H21N7O3 | 383.412 | 6 / 0 | 0.3 | Yes |
| 520264 | 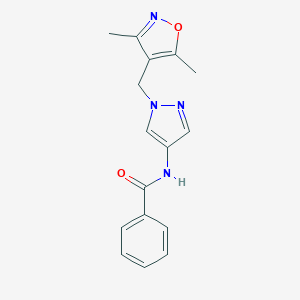 SCHEMBL1279194 SCHEMBL1279194 | C16H16N4O2 | 296.33 | 4 / 1 | 1.9 | Yes |
| 520266 | 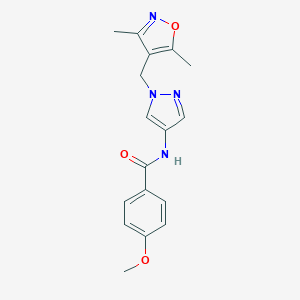 SCHEMBL1615819 SCHEMBL1615819 | C17H18N4O3 | 326.356 | 5 / 1 | 1.9 | Yes |
| 520268 | 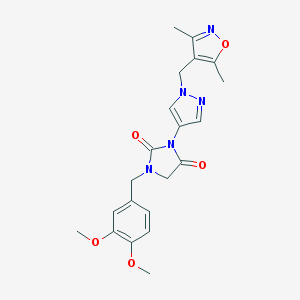 SCHEMBL1279812 SCHEMBL1279812 | C21H23N5O5 | 425.445 | 7 / 0 | 1.5 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417